I have a csv file I'm trying to read with pd.read_csv.
Some lines are just fine but other lines are grouped in the first column & the rest are filled with nan values. The problem is with those lines that have commas in the title column & are therefore grouped between quotes, like this:
Apples,Oranges,5,"These are apples, red, and oranges, orange",2;
If you open the same file with the text editor, it looks like this:
"Apples,Oranges,5,""These are apples, red, and oranges, orange"",2";
I tried a lot of different things (such as delimiter=',', quotechar='"') but nothing seems to work.
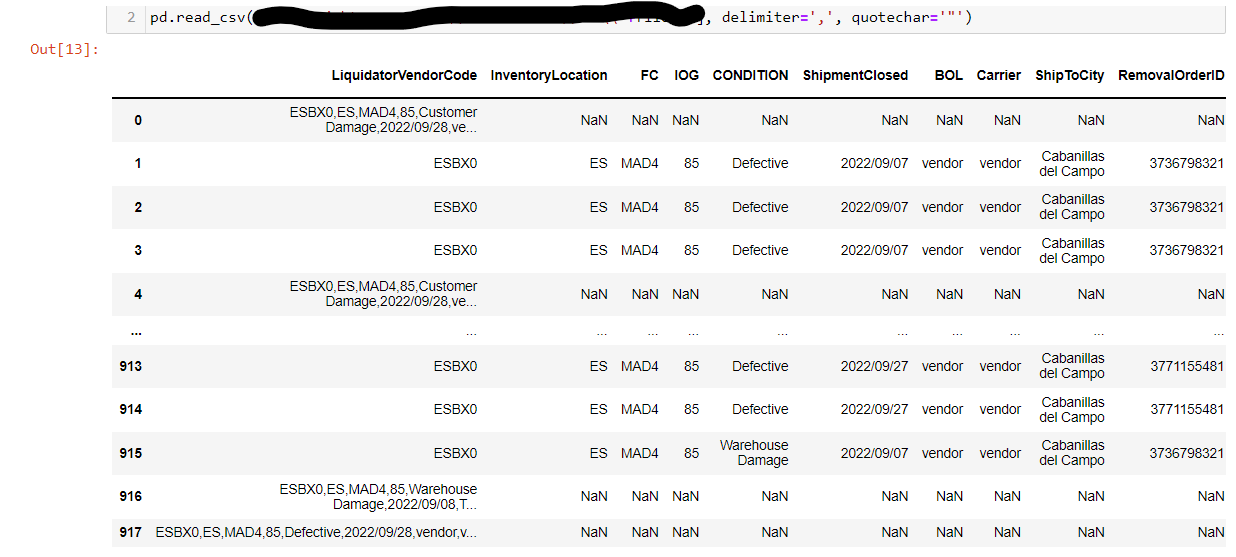
Any advice?
CodePudding user response:
Maybe you could try using something like this:
import pandas as pd
def fix_row(row: pd.Series, column_name: str) -> pd.Series:
"""Split a row into multiple rows if `column_name` is a comma separated string.
Parameters
----------
row : pd.Series
The row to split.
column_name : str
The name of the column to split.
Returns
-------
pd.Series
The original row, or row created from splitting `column_name`.
"""
value = row[column_name]
formated_value = str(value).split(',')
if len(formated_value) > 1:
return pd.Series(dict(zip(row.keys(), formated_value)))
return row
# == Example ==================================================
df = pd.DataFrame(
{
"col1": ["1234,2022-02-02,10", "1234", "EBX10", "EBX20,2022-02-02,10"],
"col2": [None, "2022-02-02", "2022-03-02", None],
"col3": [None, 10, 50, None],
}
)
# Dataframe `df` looks like this:
#
# col1 col2 col3
# 0 1234,2022-02-02,10 None NaN <-- Column with formating problem
# 1 1234 2022-02-02 10.0
# 2 EBX10 2022-03-02 50.0
# 3 EBX20,2022-02-02,10 None NaN <-- Column with formating problem
# Call `fix_row` function using apply, and specify the name of the column
# to maybe split into multiple columns.
new_df = df.apply(fix_row, column_name='col1', axis=1)
# Dataframe `new_df` looks like this:
#
# col1 col2 col3
# 0 1234 2022-02-02 10
# 1 1234 2022-02-02 10.0
# 2 EBX10 2022-03-02 50.0
# 3 EBX20 2022-02-02 10
Notes on fix_row function
The fix_row function works based on a couple of assumptions that need to be true, in order for it to work:
- The function assumes that values from
column_nameparameter (in the example above,'col1'), will only contain multiple commas, when there's a formatting problem. - When there's a row to fix, the function assumes that all the row values need to be replaced with the values from the
column_nameyou specify, and they are in the correct order.
Input and Output from the Example
Input Pandas DataFrame df:
| col1 | col2 | col3 |
|---|---|---|
| 1234,2022-02-02,10 | nan | |
| 1234 | 2022-02-02 | 10 |
| EBX10 | 2022-03-02 | 50 |
| EBX20,2022-02-02,10 | nan |
Output from df.apply(fix_row, column_name='col1', axis=1):
| col1 | col2 | col3 |
|---|---|---|
| 1234 | 2022-02-02 | 10 |
| 1234 | 2022-02-02 | 10 |
| EBX10 | 2022-03-02 | 50 |
| EBX20 | 2022-02-02 | 10 |
Variations to fix_row you might consider trying
Instead of checking the length of the formated_value, you might swap the if condition statement, checking whether the other values from the row are empty or not. You could do that using something like this:
def fix_row(row: pd.Series, column_name: str) -> pd.Series:
value = row[column_name]
formated_value = str(value).split(',')
if row[[col for col in row.keys() if col != column_name]].isna().all():
return pd.Series(dict(zip(row.keys(), formated_value)))
return row
CodePudding user response:
While doing my jogging, your question was bugging me, because of that ; you showed at the end of both your good and your bad lines. Lines in a CSV file don't usually end in a semi colon!
My theory that your file was created by a process that wrote a semi-colon-separated-value file containing in column 0 the contents of each comma-separated-value line. That process had to escape double quotes in saw in the standard way, which is the put double quotes around the field and replace each double quote within the field with a doubled double quote.
If that theory is correct, what you can do is read the file twice, once as a semi-colon-separated-value file, and then the contents of column 0 as a CSV file.
Here's code that does exactly that:
import pandas as pd
import csv
import io
buffer = io.StringIO()
with open("file.mixed-sv", newline="") as mixed_sv_file:
reader = csv.reader(mixed_sv_file, delimiter=";")
for row in reader:
print(row[0], file=buffer)
df = pd.read_csv(io.StringIO(buffer.getvalue()))
print("df:\n", df)
Given this input file, which I'm calling file.mixed-sv to highlight it's not pure CSV:
Fruit1,Fruit2,N,Notes,M;
Apples,Oranges,5,"These are apples, red, and oranges, orange",2;
"Apples,Oranges,5,""These are apples, red, and oranges, orange"",2";
my script outputs:
df:
Fruit1 Fruit2 N Notes M
0 Apples Oranges 5 These are apples, red, and oranges, orange 2
1 Apples Oranges 5 These are apples, red, and oranges, orange 2
Notes:
- I'm using the
csvmodule rather than Pandas for the initial filter, a) because I understand it better, but b) because at this stage of processing, you really don't need the power of Pandas, you just need to parse the file and extract the first field. - If your file is big, this is going to take a lot of memory because I end up with 3 copies it in memory at the same time. If you need something more memory efficient, replace my StringIO buffer by an actual temporary file on disk.
