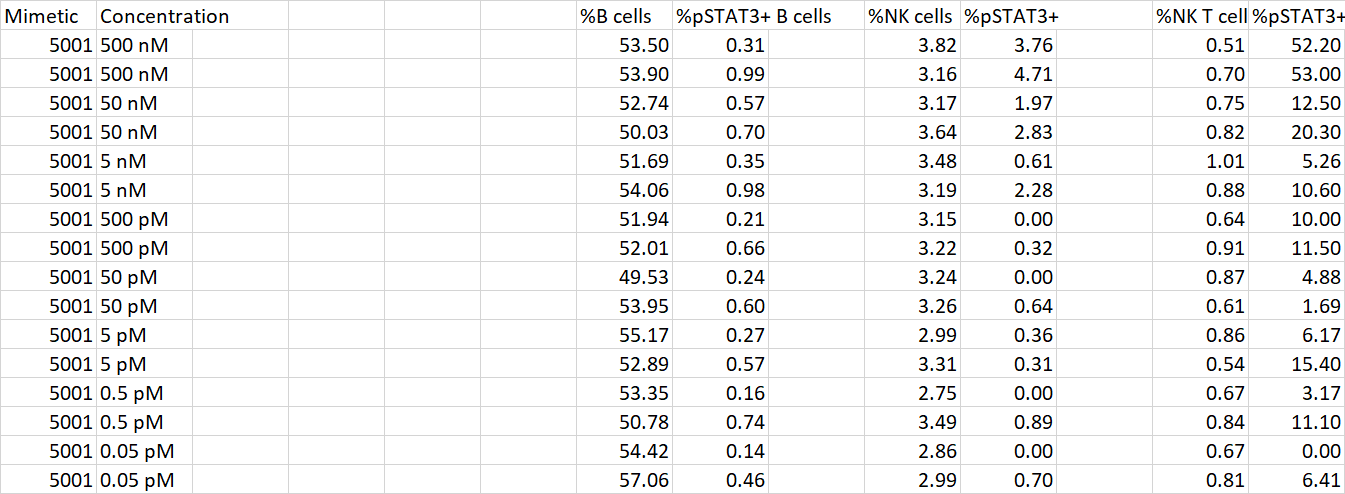
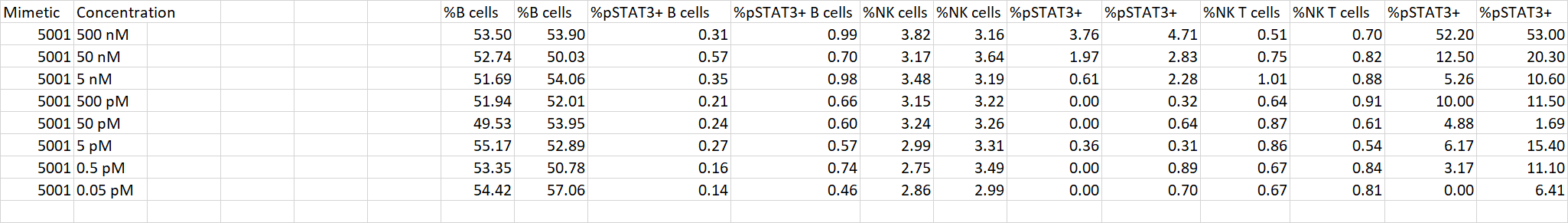
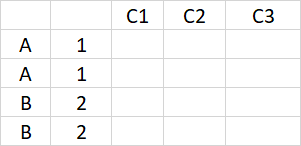
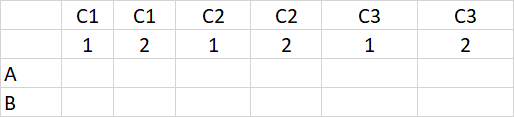
I have a data set in which each replicate of my experiment is included in an additional row, like so:
I have no idea of what approach to use in R so that next time I get data in that format, I can transform it to look like this (every replicate's column values are next to each other):
So in other words; I am trying to take
And turn it into
Can any kind human help me make this happen using R studio?
Here is the dput
structure(list(Mimetic = c("5001", "5001", "5001", "5001", "5001",
"5001", "5001", "5001", "5001", "5001", "5001", "5001", "5001",
"5001", "5001", "5001", "5002", "5002", "5002", "5002", "5002",
"5002", "5002", "5002", "5002", "5002", "5002", "5002", "5002",
"5002", "5002", "5002", "5004", "5004", "5004", "5004", "5004",
"5004", "5004", "5004", "5004", "5004", "5004", "5004", "5004",
"5004", "5004", "5004", "mIL21", "mIL21", "mIL21", "mIL21", "mIL21",
"mIL21", "mIL21", "mIL21", "mIL21", "mIL21", "mIL21", "mIL21",
"mIL21", "mIL21", "4717", "4717", "4717", "4717", "4717", "4717",
"4717", "4717", "4717", "4717", "4717", "4717", "4717", "4717",
"4717", "4717", "4934", "4934", "4934", "4934", "4934", "4934",
"4934", "4934", "4934", "4934", "4934", "4934", "4934", "4934",
"4934", "4934", "4939", "4939", "4939", "4939", "4939", "4939",
"4939", "4939", "4939", "4939", "4939", "4939", "4939", "4939",
"4939", "4939", "4943", "4943", "4943", "4943", "4943", "4943",
"4943", "4943", "4943", "4943", "4943", "4943", "4943", "4943",
"4943", "4943"), Concentration = c("500 nM", "500 nM", "50 nM",
"50 nM", "5 nM", "5 nM", "500 pM", "500 pM", "50 pM", "50 pM",
"5 pM", "5 pM", "0.5 pM", "0.5 pM", "0.05 pM", "0.05 pM", "500 nM",
"500 nM", "50 nM", "50 nM", "5 nM", "5 nM", "500 pM", "500 pM",
"50 pM", "50 pM", "5 pM", "5 pM", "0.5 pM", "0.5 pM", "0.05 pM",
"0.05 pM", "500 nM", "500 nM", "50 nM", "50 nM", "5 nM", "5 nM",
"500 pM", "500 pM", "50 pM", "50 pM", "5 pM", "5 pM", "0.5 pM",
"0.5 pM", "0.05 pM", "0.05 pM", "5 nM", "5 nM", "500 pM", "500 pM",
"50 pM", "50 pM", "5 pM", "5 pM", "0.5 pM", "0.5 pM", "0.05 pM",
"0.05 pM", "0", "0", "500 nM", "500 nM", "50 nM", "50 nM", "5 nM",
"5 nM", "500 pM", "500 pM", "50 pM", "50 pM", "5 pM", "5 pM",
"0.5 pM", "0.5 pM", "0.05 pM", "0.05 pM", "500 nM", "500 nM",
"50 nM", "50 nM", "5 nM", "5 nM", "500 pM", "500 pM", "50 pM",
"50 pM", "5 pM", "5 pM", "0.5 pM", "0.5 pM", "0.05 pM", "0.05 pM",
"500 nM", "500 nM", "50 nM", "50 nM", "5 nM", "5 nM", "500 pM",
"500 pM", "50 pM", "50 pM", "5 pM", "5 pM", "0.5 pM", "0.5 pM",
"0.05 pM", "0.05 pM", "500 nM", "500 nM", "50 nM", "50 nM", "5 nM",
"5 nM", "500 pM", "500 pM", "50 pM", "50 pM", "5 pM", "5 pM",
"0.5 pM", "0.5 pM", "0.05 pM", "0.05 pM"), X.B.cells = c(53.5,
53.9, 52.74, 50.03, 51.69, 54.06, 51.94, 52.01, 49.53, 53.95,
55.17, 52.89, 53.35, 50.78, 54.42, 57.06, 56.58, 54.81, 55.29,
55.69, 57.4, 56.04, 59.18, 60.57, 60.46, 55.85, 66.27, 61.11,
64.85, 59.97, 72.06, 70.51, 53.4, 55.78, 58.22, 53.57, 59.61,
56.68, 58.99, 53.9, 60.49, 58.24, 63.7, 62.77, 61.19, 59.18,
65.81, 58.18, 51.3, 50.38, 58.22, 48.96, 49.38, 59.08, 54.34,
50.19, 52.68, 50.86, 52.12, 48.53, 68.55, 55.61, 62.96, 62.69,
65.3, 60.66, 57.37, 58.27, 56.37, 56.8, 59.11, 55.13, 63.47,
65.06, 68.19, 67, 56.9, 54.98, 61.17, 60.67, 60.35, 64.51, 60.85,
63.46, 62.69, 62.12, 61.65, 58.97, 65.61, 67.04, 63.44, 69.4,
66.35, 67.79, 60.95, 66.13, 62.99, 64.5, 64.41, 63.28, 58.33,
64.74, 63.52, 61.12, 62.24, 67.12, 64.82, 69.05, 65.07, 67.8,
59.98, 61.03, 59.49, 58.11, 58.63, 54.01, 58.64, 59.09, 53.08,
54.53, 55.8, 54.82, 51.04, 59.63, 58.19, 60.26), X.pSTAT3..B.cells = c(0.31,
0.99, 0.57, 0.7, 0.35, 0.98, 0.21, 0.66, 0.24, 0.6, 0.27, 0.57,
0.16, 0.74, 0.14, 0.46, 0.86, 0.92, 0.19, 0.41, 0.4, 0.22, 0.07,
0.1, 0.07, 0.08, 0.16, 0.27, 0.46, 0.21, 0.21, 0.09, 4.03, 3.45,
2.06, 2.54, 1.26, 1.15, 0.18, 0.2, 0.21, 0.36, 0.07, 0.17, 0.1,
0.32, 0.35, 0.25, 1.66, 2.3, 4.31, 5.19, 4.16, 4.31, 3.31, 3.94,
1.4, 2.47, 0.23, 1.17, 0.5, 0.46, 8.46, 14.4, 8.19, 13.4, 2.93,
5.24, 0.95, 2.81, 0.39, 1, 0.29, 0.94, 0.34, 0.31, 0.55, 0.72,
8.35, 8.85, 3.5, 2.77, 1.71, 1.33, 1.94, 1.42, 0.67, 0.87, 0.72,
0.77, 0.49, 0.48, 0.25, 0.49, 2.49, 1.57, 1.18, 3.05, 0.9, 2.13,
1.16, 1.44, 0.75, 1.23, 0.53, 1.02, 1.69, 0.81, 0.22, 0.46, 6.12,
9.88, 6.84, 8.08, 2.93, 2.01, 1.75, 1.09, 0.39, 0.8, 0.96, 0.72,
0.21, 0.39, 0.58, 0.84), X.NK.cells = c(3.82, 3.16, 3.17, 3.64,
3.48, 3.19, 3.15, 3.22, 3.24, 3.26, 2.99, 3.31, 2.75, 3.49, 2.86,
2.99, 3.1, 2.96, 3.01, 3.16, 2.52, 2.74, 1.99, 2.49, 2.72, 3.06,
1.6, 2.53, 1.57, 2.45, 1.21, 2.09, 3.33, 2.66, 2.48, 2.92, 2.17,
3.01, 2.25, 3.44, 2.18, 2.94, 2.04, 2.25, 2.11, 2.57, 2.18, 2.92,
2.73, 3.31, 2.35, 3.6, 3.92, 2.45, 3.19, 3.31, 3.35, 3.25, 3.39,
3.91, 2.1, 3.4, 2.69, 2.1, 2.54, 2.7, 3.17, 2.91, 2.98, 2.6,
2.53, 3.07, 2.04, 1.6, 1.25, 1.69, 2.39, 3.19, 2.47, 2.16, 3.01,
1.72, 2.37, 2.12, 2.39, 2.71, 2.1, 2.85, 2.17, 1.63, 2.17, 1.45,
1.85, 1.7, 2.41, 1.69, 2.9, 1.82, 2.27, 2.3, 3.15, 1.86, 2.29,
2.55, 2.34, 1.93, 2.2, 1.86, 1.93, 1.66, 3.46, 2.24, 3.02, 3.07,
3.1, 3.26, 3.29, 2.34, 3.03, 3.16, 3.23, 3.18, 3.08, 2.77, 2.97,
2.56), X.pSTAT3. = c(3.76, 4.71, 1.97, 2.83, 0.61, 2.28, 0, 0.32,
0, 0.64, 0.36, 0.31, 0, 0.89, 0, 0.7, 2.78, 1.41, 0.69, 3.27,
1.65, 0, 0, 2.5, 1.53, 0.68, 2.63, 0, 0, 0, 0, 0, 6.27, 5.53,
4.17, 2.11, 2.88, 6.29, 0.93, 0.61, 0, 0, 0, 0, 0, 0, 0.97, 0,
0, 2.41, 8.38, 6.74, 2.58, 1.4, 3.64, 1.6, 0.51, 2.93, 0.36,
1.09, 1.82, 3.11, 11.5, 17.6, 12.2, 17.6, 5.23, 7.69, 2.08, 7.09,
0.82, 4, 0, 1.28, 0, 2.44, 0, 0, 7.56, 13.3, 8.84, 9.52, 6.03,
1.94, 6.9, 5.3, 5.88, 3.62, 0, 1.27, 0.95, 0, 2.25, 0, 9.48,
4.94, 2.86, 12.5, 0.91, 6.31, 1.31, 3.33, 0, 2.46, 1.77, 3.23,
3.77, 1.11, 0, 1.25, 13.9, 18.5, 8.84, 12, 5.33, 7.55, 3.82,
1.77, 1.36, 2.61, 1.28, 1.3, 0, 2.27, 2.84, 2.46), X.NK.T.cells = c(0.51,
0.7, 0.75, 0.82, 1.01, 0.88, 0.64, 0.91, 0.87, 0.61, 0.86, 0.54,
0.67, 0.84, 0.67, 0.81, 0.72, 0.65, 0.6, 0.68, 0.96, 0.64, 0.62,
0.6, 0.69, 0.75, 0.72, 0.52, 0.78, 0.72, 0.72, 0.68, 0.79, 0.83,
0.83, 0.56, 0.83, 1.16, 0.82, 0.85, 0.67, 0.75, 0.62, 0.84, 0.77,
0.55, 0.63, 0.89, 0.43, 1.02, 1.04, 0.69, 1.27, 0.58, 0.8, 0.75,
1.15, 1, 0.56, 1.19, 0.5, 0.66, 0.6, 0.51, 0.5, 0.82, 0.95, 0.94,
0.85, 0.7, 1.1, 0.88, 0.69, 0.65, 0.92, 0.47, 1.15, 0.87, 0.58,
0.56, 0.41, 0.6, 0.72, 0.41, 0.87, 0.37, 0.66, 0.83, 0.6, 0.41,
0.35, 0.52, 0.62, 0.66, 0.73, 0.46, 0.62, 0.68, 0.85, 0.58, 0.72,
0.54, 0.52, 0.63, 0.56, 0.52, 0.56, 0.45, 0.37, 0.73, 0.92, 0.73,
1.27, 0.7, 1.9, 0.72, 1.7, 0.85, 1.2, 0.62, 0.99, 0.76, 1.26,
1.24, 1.56, 0.86), X.pSTAT3..1 = c(52.2, 53, 12.5, 20.3, 5.26,
10.6, 10, 11.5, 4.88, 1.69, 6.17, 15.4, 3.17, 11.1, 0, 6.41,
20.9, 16.1, 3.45, 15.2, 13, 6.45, 3.33, 3.45, 12.1, 0, 5.88,
0, 2.7, 8.82, 11.8, 0, 47.4, 38, 15, 22.2, 15, 18.2, 5.13, 10,
6.25, 8.33, 3.45, 7.5, 8.11, 3.85, 6.67, 7.14, 41.2, 31.3, 57,
55.9, 38.6, 54.9, 52.7, 35.2, 16.4, 16.8, 0, 10.8, 7.69, 11.4,
75.9, 76, 83.3, 87.5, 52.2, 54.3, 17.1, 38.2, 9.43, 23.3, 6.06,
9.38, 13.6, 0, 12.7, 14.3, 64.3, 55.6, 30, 37.9, 28.6, 20, 38.1,
16.7, 9.38, 5, 3.45, 15, 17.6, 4, 6.67, 6.25, 60, 50, 26.7, 30.3,
12.2, 32.1, 25.7, 30.8, 20, 3.33, 3.7, 12, 18.5, 9.09, 11.1,
5.71, 63.6, 65.7, 62.9, 58.8, 32.6, 48.6, 13.6, 14.6, 12.1, 10,
8.33, 10.8, 6.67, 6.78, 2.7, 7.32), X.CD4..T.cells = c(17.9,
18.39, 19.12, 20.22, 20.15, 19.68, 20.96, 20.04, 21.49, 19.71,
18.93, 20.23, 20.25, 19.83, 19.62, 18.36, 17.85, 19.5, 18.87,
19.33, 18.15, 19.43, 18.8, 18.08, 17.74, 19.49, 17.41, 18.63,
18.65, 18.78, 14.53, 13.87, 19.57, 19.11, 19.31, 20.51, 19.51,
20.4, 19.09, 19.48, 18.12, 18.15, 16.61, 16.96, 18.52, 18.24,
15.59, 19.45, 23.08, 21.97, 18.36, 20.96, 21.24, 18.68, 20.23,
21.54, 21.34, 21.33, 21.14, 22.03, 15.03, 19.23, 16.33, 16.16,
15.1, 16.47, 18.23, 17.52, 17.74, 18.45, 17.07, 18.18, 17.85,
16.63, 15.25, 16, 19.36, 18.82, 16.65, 16.89, 17.13, 15.93, 16.11,
16.87, 15.99, 16.19, 18.18, 16.75, 15.87, 14.86, 16.36, 14.88,
16.22, 14.69, 16.44, 15.31, 16.21, 14.6, 14.84, 17.05, 16.58,
16.05, 16.53, 15.5, 17.37, 14.12, 15.01, 14.43, 15.81, 14.35,
16.28, 17.46, 16.91, 18.18, 17.73, 19.25, 18.07, 17.96, 20.03,
19.17, 18.59, 19.29, 20.42, 17.72, 17.37, 17.8), X.pSTAT3..2 = c(5.85,
7.63, 0.98, 0.82, 0.42, 1.22, 0.31, 1.04, 0.54, 1, 0.28, 0.92,
0.21, 0.52, 0.11, 0.23, 5.01, 3.75, 1.54, 1.28, 0.57, 1.5, 0.44,
0.8, 0.35, 0.32, 0.97, 0.11, 0.45, 0.45, 0.29, 0.31, 12.6, 10.7,
8.35, 8.03, 4.92, 5.06, 3.08, 1.3, 0.81, 0.57, 0.77, 0.62, 0.34,
0.23, 0.41, 0.65, 6.09, 5.56, 5.22, 16.1, 5.79, 10.9, 5.16, 8.69,
1.36, 3.72, 0.75, 1.02, 1.02, 1.73, 26, 34.2, 26.4, 37, 10.9,
16.8, 1.28, 4.55, 0.85, 2.47, 0.58, 0.98, 2.19, 2.06, 0.11, 0.55,
23.6, 27, 7.18, 8.51, 4.57, 4.51, 4.38, 3.04, 1.7, 2.34, 1.56,
1.66, 1.77, 1.25, 0.51, 1.27, 16.3, 9.92, 4.34, 8.24, 1.67, 4.74,
2.61, 3.09, 1.62, 2.02, 1.43, 1.62, 4.97, 1.15, 0.52, 0.43, 20.7,
30, 19.6, 24.8, 5.25, 6.5, 1.62, 1.38, 0.41, 0.65, 0.56, 1.18,
0.31, 0.95, 0.36, 0.12), X.Tregs = c(0.86, 0.86, 1.01, 0.67,
0.9, 0.7, 0.75, 0.74, 1.07, 0.8, 0.81, 0.73, 0.65, 0.68, 0.67,
0.68, 0.86, 0.82, 0.54, 0.47, 0.73, 0.58, 0.5, 0.64, 0.73, 0.71,
0.78, 0.88, 0.8, 0.63, 0.7, 0.66, 0.87, 1.05, 0.72, 0.91, 0.79,
1.09, 0.74, 0.76, 0.69, 0.69, 0.55, 0.48, 0.63, 0.72, 0.68, 1,
1.83, 1.23, 1.3, 1.2, 1.53, 0.89, 1.44, 0.77, 1.44, 1.02, 0.77,
1.31, 0.57, 0.97, 0.56, 0.8, 0.66, 1.17, 0.85, 1.02, 0.5, 0.57,
0.83, 1.04, 0.75, 0.76, 0.71, 0.87, 0.55, 1.06, 0.77, 0.6, 0.84,
0.76, 0.78, 0.6, 0.8, 0.78, 0.97, 0.76, 0.56, 0.82, 0.62, 0.6,
0.91, 0.89, 0.94, 0.69, 0.87, 0.6, 0.58, 1.16, 0.53, 0.64, 0.76,
0.69, 0.64, 0.79, 0.66, 0.54, 0.77, 0.77, 0.77, 0.5, 0.76, 0.74,
1.26, 0.82, 1.09, 0.83, 1.13, 0.83, 1.1, 0.81, 0.63, 0.69, 1.03,
0.76), X.pSTAT3..3 = c(38.5, 51.9, 4.12, 9.23, 0, 2.99, 2.86,
4.23, 0, 2.6, 1.32, 4.29, 0, 0, 0, 1.54, 23.8, 21.8, 7.69, 13,
5.71, 0, 0, 0, 0, 0, 0, 2.38, 2.63, 0, 0, 0, 31.3, 31, 11.4,
27.3, 21.1, 28.8, 17.1, 13.9, 0, 9.09, 0, 0, 3.33, 5.88, 3.12,
2.13, 9.59, 20.1, 19.2, 31.1, 15.1, 21.8, 16.2, 21.9, 5.95, 12.5,
3.17, 3.25, 0, 3.12, 59.3, 56.4, 43.8, 59.6, 22, 32, 4.17, 7.14,
5, 7.84, 5.56, 2.7, 5.88, 4.76, 0, 0, 51.4, 72.4, 17.1, 18.9,
10.5, 34.5, 15.4, 10.5, 8.51, 10.8, 3.7, 2.5, 10, 6.9, 0, 2.33,
51.1, 63.6, 26.2, 34.5, 3.57, 21.4, 15.4, 3.23, 10.8, 9.09, 6.45,
5.26, 18.8, 7.69, 0, 2.7, 59.5, 58.3, 40.5, 47.2, 13.1, 17.5,
9.62, 5, 0, 7.5, 1.89, 2.56, 3.33, 0, 2.04, 0), X.CD8..T.cells = c(19.88,
18.88, 18.27, 18.9, 18.74, 17.72, 19.17, 19.09, 20.3, 18.09,
18.23, 18.56, 18.15, 20.19, 18.41, 16.41, 17.73, 17.9, 16.82,
15.14, 17.61, 16.52, 15.43, 14.24, 14.62, 15.6, 10.89, 13.1,
11.48, 13.48, 8.86, 9.89, 19.02, 18.2, 15.11, 16.37, 14.33, 14.94,
15.24, 17.49, 14.6, 15.01, 13.37, 13.4, 13.61, 15.49, 12.54,
14.49, 20.27, 19.73, 16.23, 21.14, 18.88, 15.27, 17.39, 19.84,
16.23, 19.64, 18.5, 20.83, 10.94, 17.45, 12.35, 11.7, 11.4, 12.63,
16.52, 14.55, 17.7, 15.83, 15.85, 17.44, 12.32, 11.35, 11.09,
10.59, 16.99, 17.37, 13.29, 12.55, 12.75, 9.28, 12.51, 10.66,
12.77, 12.17, 12.46, 13.24, 10.79, 9.38, 11.35, 8.19, 10.47,
9.78, 13.09, 11.22, 11.55, 11.88, 11.18, 12.02, 13.54, 11.62,
11.54, 13.33, 11.78, 10.37, 12.71, 9.61, 10.67, 10.57, 15.19,
13.94, 15.74, 14.86, 14.77, 17.77, 14.99, 15.54, 18.81, 18.57,
17.93, 17.76, 20.56, 14.66, 16.72, 15.32), X.pSTAT3..4 = c(12.7,
12.9, 0.63, 1.47, 0.17, 0.7, 0.17, 0.87, 0.21, 0.8, 0, 0.56,
0.12, 0.62, 0.06, 0.32, 9.9, 6.25, 1.23, 1.63, 0.59, 0.75, 0.13,
0.29, 0.28, 0, 0.19, 0.16, 0, 0.16, 0.48, 0, 23.4, 18.6, 17.2,
16.1, 10.3, 6.9, 0.55, 1.57, 0.29, 0.14, 0.63, 0.16, 0.31, 0.27,
0.17, 0, 14.9, 12.1, 13.3, 29.4, 13.2, 20.2, 12.1, 16.4, 2.42,
6.73, 0.13, 0.77, 0.7, 1.13, 43.1, 53.6, 42.4, 62.8, 28, 37.9,
2.92, 8.93, 0.52, 1.29, 0.51, 0.72, 0.56, 0.78, 0.37, 0.48, 40.3,
48.5, 15.1, 18.8, 2.78, 5.6, 4.35, 3.04, 1.49, 1.25, 1.15, 1.1,
0, 0, 0.4, 0.21, 34.3, 17.4, 4.12, 11, 1.29, 5.86, 1.67, 2.14,
1.25, 0.94, 0.53, 0.6, 2.93, 1.08, 0, 0, 30.2, 48.1, 39.2, 45.1,
15.4, 17.8, 2.38, 2, 0.22, 0.56, 0.46, 0.93, 0.2, 0.43, 0.13,
0.55), X.TCRgd.T.cells = c(0.64, 0.6, 0.55, 0.51, 0.39, 0.54,
0.48, 0.49, 0.64, 0.51, 0.55, 0.54, 0.55, 0.59, 0.42, 0.54, 0.45,
0.43, 0.41, 0.39, 0.44, 0.35, 0.6, 0.5, 0.31, 0.4, 0.4, 0.23,
0.49, 0.38, 0.36, 0.34, 0.67, 0.47, 0.43, 0.6, 0.35, 0.36, 0.53,
0.51, 0.5, 0.52, 0.25, 0.38, 0.35, 0.42, 0.32, 0.61, 0.55, 0.64,
0.53, 0.56, 0.55, 0.41, 0.39, 0.49, 0.41, 0.34, 0.41, 0.75, 0.38,
0.36, 0.44, 0.25, 0.21, 0.39, 0.37, 0.43, 0.48, 0.43, 0.37, 0.41,
0.35, 0.29, 0.31, 0.45, 0.5, 0.41, 0.19, 0.47, 0.35, 0.33, 0.14,
0.27, 0.37, 0.39, 0.27, 0.29, 0.14, 0.33, 0.5, 0.29, 0.21, 0.31,
0.48, 0.29, 0.39, 0.37, 0.25, 0.23, 0.47, 0.27, 0.31, 0.29, 0.46,
0.25, 0.33, 0.27, 0.19, 0.29, 0.27, 0.35, 0.31, 0.41, 0.35, 0.47,
0.42, 0.5, 0.35, 0.54, 0.46, 0.43, 0.63, 0.42, 0.5, 0.21), X.pSTAT3..5 = c(15.5,
19.6, 3.77, 2.04, 0, 1.92, 0, 4.26, 0, 0, 0, 0, 0, 0, 0, 0, 11.9,
9.76, 5, 5.26, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 26.6, 13.3,
4.76, 13.8, 0, 11.8, 0, 12.5, 4.17, 0, 0, 5.56, 0, 0, 0, 0, 0,
6.94, 15, 21.8, 2.63, 19.4, 3.7, 4.35, 0, 3.12, 0, 0, 0, 4.17,
42.9, 58.3, 30, 42.1, 11.1, 38.1, 4.35, 0, 0, 0, 0, 0, 0, 0,
0, 0, 77.8, 34.8, 5.88, 0, 14.3, 7.69, 5.56, 0, 7.69, 0, 0, 0,
4.17, 7.14, 0, 0, 52.2, 21.4, 10.5, 5.56, 0, 0, 0, 0, 0, 0, 0,
0, 0, 7.69, 0, 0, 23.1, 47.1, 20, 25, 11.8, 8.7, 5, 0, 0, 0,
0, 0, 0, 0, 0, 0)), class = "data.frame", row.names = c(NA, -126L
))
CodePudding user response:
I think this works too:
library(tidyverse)
df %>%
group_by(Mimetic, Concentration) %>%
mutate(obs = row_number()) %>%
ungroup() %>%
pivot_wider(names_from = c(Concentration, obs), values_from = 3:16)
We can use glimpse to see what the very wide output looks like:
Rows: 8
Columns: 253
$ Mimetic <chr> "5001", "5002", "5004", "mIL21", "4717", "4934", "4939", "4943"
$ `X.B.cells_500 nM_1` <dbl> 53.50, 56.58, 53.40, NA, 62.96, 61.17, 60.95, 59.98
$ `X.B.cells_500 nM_2` <dbl> 53.90, 54.81, 55.78, NA, 62.69, 60.67, 66.13, 61.03
$ `X.B.cells_50 nM_1` <dbl> 52.74, 55.29, 58.22, NA, 65.30, 60.35, 62.99, 59.49
$ `X.B.cells_50 nM_2` <dbl> 50.03, 55.69, 53.57, NA, 60.66, 64.51, 64.50, 58.11
$ `X.B.cells_5 nM_1` <dbl> 51.69, 57.40, 59.61, 51.30, 57.37, 60.85, 64.41, 58.63
$ `X.B.cells_5 nM_2` <dbl> 54.06, 56.04, 56.68, 50.38, 58.27, 63.46, 63.28, 54.01
$ `X.B.cells_500 pM_1` <dbl> 51.94, 59.18, 58.99, 58.22, 56.37, 62.69, 58.33, 58.64
$ `X.B.cells_500 pM_2` <dbl> 52.01, 60.57, 53.90, 48.96, 56.80, 62.12, 64.74, 59.09
$ `X.B.cells_50 pM_1` <dbl> 49.53, 60.46, 60.49, 49.38, 59.11, 61.65, 63.52, 53.08
etc.
CodePudding user response:
This should do the trick:
library(tidyverse)
df <- df %>%
group_by(Mimetic, Concentration) %>%
summarise(across(everything(), ~list(.))) %>%
unnest_wider(everything(), names_sep = "_")
The unnest_wider function splits the nested list in a wide fashion, yielding the expected results.