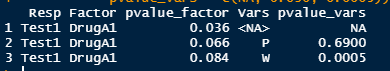
I have an output from a model which looks like the Test variable from the following code:
Test <- vector("list" ,2)
names(Test) <- c("Test1", "Test 2")
Test$Test1$factor <- c("Drug1" = 0.036)
Test$Test1$P <- c("Drug1" = 0.066, "P" = 0.69)
Test$Test1$W <- c("Drug1" = 0.084, "W" = 0.0005)
Test$Test2$factor <- c("Drug2" = 0.036)
Test$Test2$P <- c("Drug2" = 0.096, "P" = 0.069)
Test$Test2$W <- c("Drug2" = 0.046, "W" = 0.0105)
Test
However, I want the final output to be a dataframe for each of the tests that looks like this:
data.frame("Resp" = rep("Test1",length(Test$Test1)), Factor = rep("DrugA1",length(Test$Test1)),
pvalue_factor = c( 0.036, 0.066, 0.084), Vars = c(NA, names(Test$Test1)[-1]),
pvalue_vars = c(NA, 0.690, 0.0005))
and similar tabel for Test2
CodePudding user response:
I would do it like this:
# remove the extra "Test 2" item
Test <- Test[names(Test) != 'Test 2']
# table for each test
test.tab <- lapply(Test, function(tst) {
tab <- t(sapply(tst, '[', 1:2))
tab <- data.frame(Factor=colnames(tab)[1], pvalue_factor=tab[, 1],
Vars=rownames(tab), pvalue_vars=tab[, 2])
tab$Vars[tab$Vars=='factor'] <- NA
tab
})
# combine individual tables
df <- do.call(rbind, test.tab)
# add response
df <- cbind(Resp=rep(names(test.tab), sapply(test.tab, nrow)),
df)
rownames(df) <- NULL
df
# Resp Factor pvalue_factor Vars pvalue_vars
# 1 Test1 Drug1 0.036 <NA> NA
# 2 Test1 Drug1 0.066 P 0.6900
# 3 Test1 Drug1 0.084 W 0.0005
# 4 Test2 Drug2 0.036 <NA> NA
# 5 Test2 Drug2 0.096 P 0.0690
# 6 Test2 Drug2 0.046 W 0.0105