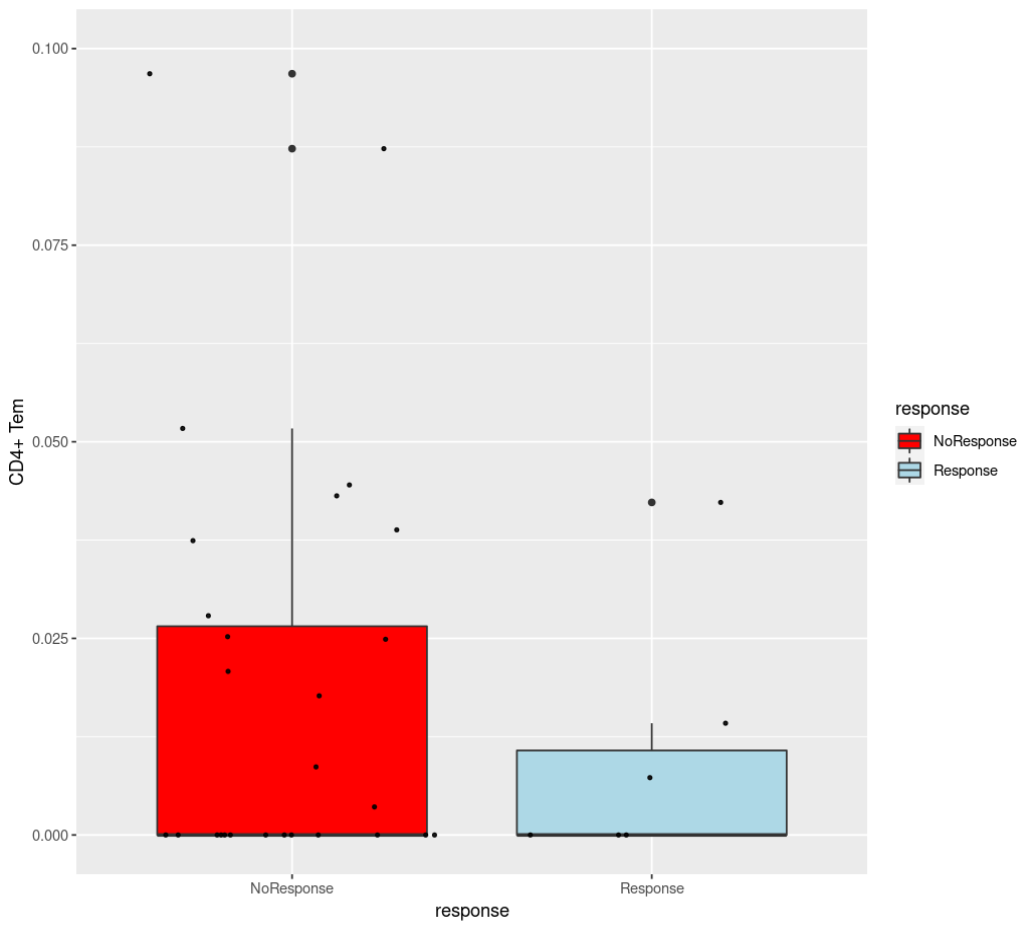
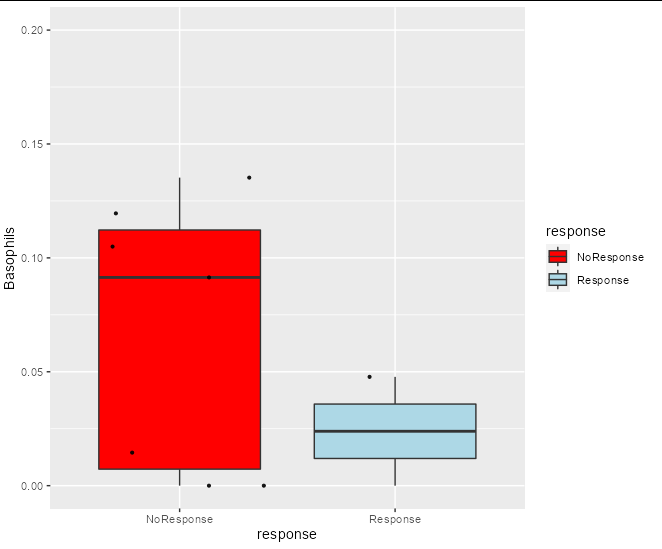
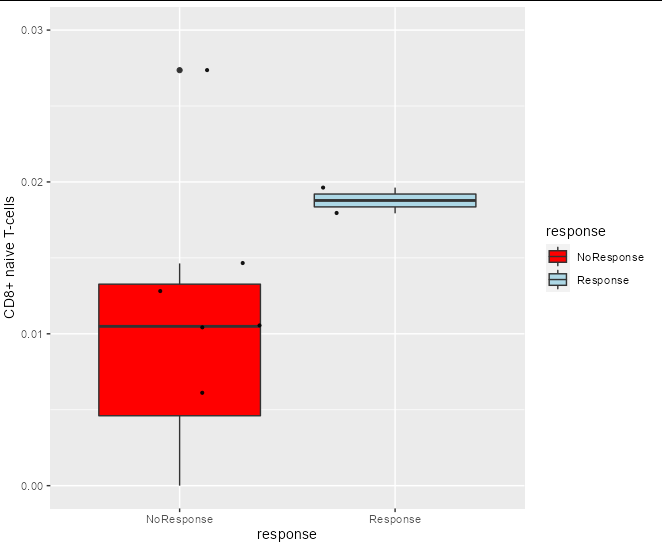
I want to make a function that presents boxplots of some data. The code works only when it's not in a function. When I put it in a function it fails. I want to put it in a function because I have many datasets of the same structure and I don't want to repeat the same code. Here is the regular code and the plot is produced:
resp <- data.frame(sample = colnames(scores), response = metadata$Benefit)
scores %>%
t() %>%
as.data.frame() %>%
rownames_to_column("sample") %>%
right_join(resp, .) %>%
ggplot(aes(x = response, y = `CD4 Tem`))
geom_boxplot(aes(fill = response))
scale_fill_manual(values = c('red','lightblue'))
geom_jitter(color="black", size=0.8, alpha=0.9)
ylim(0, 0.1)
In a function, however, it does not recognize the y = CD4 Tem argument:
Boxes <- function(scores_data ,meta_data, column, celltype, k) { ## gg boxplots
resp = data.frame(sample = colnames(scores_data), response = meta_data[[column]])
scores_data %>% t() %>% as.data.frame() %>%
rownames_to_column("sample") %>%
right_join(resp, .) %>%
ggplot(aes(x = response, y = celltype))
geom_boxplot(aes(fill = response))
scale_fill_manual(values = c('red','lightblue'))
geom_jitter(color="black", size=0.8, alpha=0.9)
ylim(0, k)
}
for example: Boxes(scores, metadata, 'Benefit', 'Tgd cells' , 0.2 )
produces:
Error in FUN(X[[i]], ...) : object 'Tgd cells' not found
Sample of scores:
structure(c(0.00175333630321122, 0.0160232697957133, 0.119533527060973,
3.38920032450346e-20, 0.00577191441771172, 1.43215113814464e-19,
0.0487323510727856, 0.0872735021313101, 0, 0.0066498486266948,
0.000940199906083161, 6.29830164958833e-20, 8.70346113353383e-19,
0.000143134161959237, 2.96379130529708e-18, 2.79878821634228e-19,
0.0303662173906541, 0, 0.0105316765508914, 0.00150757125433767,
0, 2.96649645737511e-21, 0.0477634940125643, 0.00470920090451904,
5.42614855792624e-20, 4.57868960693047e-20, 4.10374452367781e-19,
0, 0.017933442808207, 0, 0, 0.204175059886713, 0.104955515405957,
0, 0.0637468109054434, 0.0134166264055911, 0.0290285592482964,
0.0516962126912615, 0.00613804602680461, 0.0710305792589827,
0.00778137172189777, 0.000125708674793805, 0.353609047272269,
1.512434582877e-19, 3.37240641744559e-18, 2.21761495132341e-18,
0.0107824997605205, 0, 0.0128134041327042, 2.41313695610163e-18,
0.0730722604536895, 0.041888143774871, 1.48680732530708e-18,
0.0371111549542145, 0, 0.0147282080390037, 0, 2.57491956764445e-21,
0.0104516046565067, 0.0433162672733649, 8.15084122983224e-21,
0.0380473033612007, 0.0145099994894181, 0.014147791345196, 2.71228259849911e-18,
0.0155558910245464, 2.21377704157198e-18, 2.87358596630636e-19,
0.0146348072134295, 0, 0.00139414029448817, 0, 0.0914092782821773,
0.00870970590818875, 0, 0, 3.67839239412613e-19, 0, 0.0273601871652021,
7.93119212240358e-20, 0, 0.0296730264184565, 0, 0.0215283348860577,
4.08767873819977e-18, 0.00247702696715108, 2.81027335370524e-18,
0, 0.0196274656724322, 0, 0.00181974101325591, 0.0209131793129819,
0.135232905818028, 0.0182668507516641, 2.96556952666363e-18,
4.90745252242977e-21, 0.0608518971538323, 0, 0, 0), dim = c(10L,
10L), dimnames = list(c("Adipocytes", "B-cells", "Basophils",
"CD4 memory T-cells", "CD4 naive T-cells", "CD4 T-cells",
"CD4 Tcm", "CD4 Tem", "CD8 naive T-cells", "CD8 T-cells"),
c("Pat02", "Pat03", "Pat04", "Pat06", "Pat08", "Pat118",
"Pat119", "Pat123", "Pat126", "Pat14")))
sample of metadata:
structure(list(Benefit = c("NoResponse", "NoResponse", "Response",
"NoResponse", "NoResponse", "NoResponse", "NoResponse", "NoResponse",
"Response", "NoResponse"), age_start = c(42, 61, 71, 33, 73,
43, 61, 50, 77, 48), gct = c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1), RECIST = c("SD",
"PD", "PR", "PD", "PD", "PD", "PD", "SD", "PR", "PD"), overall_survival = c(1632,
100, 987, 161, 140, 313, 808, 853, 641, 163), progression_free = c(538,
76, 646, 77, 73, 120, 108, 544, 187, 84), primary = c("occult",
"skin", "skin", "skin", "mucosal", "skin", "skin", "occult",
"skin", "skin"), group = c("response", "nonresponse", "response",
"nonresponse", "nonresponse", "nonresponse", "long-survival",
"response", "response", "nonresponse"), histology = c("Unknown or NA",
"Acrolentigenous", "Nodular", "Superficial spreading", "Nodular",
"Unknown or NA", "Unknown or NA", "Unknown or NA", "Nodular",
"Acrolentigenous"), stage = c("Stage 4", "Stage 4", "Stage 4",
"Stage 4", "Stage 4", "Stage 4", "Stage 3", "Stage 4", "Stage 4",
"Stage 4")), row.names = c("Pat02", "Pat03", "Pat04", "Pat06",
"Pat08", "Pat118", "Pat119", "Pat123", "Pat126", "Pat14"), class = "data.frame")
CodePudding user response:
You have to deal with non-standard evaluation when passing column names to a function that uses ggplot. The ideal way to do this is to use curly-curly notation {{variable}} inside the aes call. This allows you to pass column names without having to quote them, tidyverse-style:
Boxes <- function(scores_data ,meta_data, column, celltype, k) {
resp = data.frame(sample = colnames(scores_data),
response = meta_data[[deparse(substitute(column))]])
scores_data %>%
t() %>%
as.data.frame() %>%
rownames_to_column("sample") %>%
right_join(resp, .) %>%
ggplot(aes(x = response, y = {{celltype}}))
geom_boxplot(aes(fill = response))
scale_fill_manual(values = c('red','lightblue'))
geom_jitter(color="black", size=0.8, alpha=0.9)
ylim(0, k)
}
This allows
Boxes(scores, metadata, Benefit, Basophils , 0.2)
and
Boxes(scores, metadata, Benefit, `CD8 naive T-cells` , 0.03)