I am adding two different csv columns to make 2D hist plot. I have two different types of data which are dense and collision.
Further, each data includes information of my case study with column type where I have type=0 (big) and type=1(small).
csv looks like this(from collision):
TIMESTEP id type a |f| |v|
20000 4737 0 9.81 1.31495 4.18007
40000 11991 1 9.81 4.43794 4.17909
50000 15725 1 9.81 4.43794 4.17810
30000 8209 0 9.81 4.43794 4.17810
15000 3545 0 9.81 1.31495 4.17810
30000 8269 0 9.81 4.43794 4.17810
10000 2077 1 9.81 1.31495 4.17712
20000 5079 0 9.81 1.31495 4.17712
All data are float types with positive entries.
When I plot a and |f| from both types for type=0 (big) and type=1(small) separetely I don't have any problem. Also plot makes sense. However, plotting a and |f| from small big (i.e sum of each entry from each part) looks weird.
I realized small big gave me almost 90% Nan though original data doesn't have any Nan.
How can I avoid Nan while doing small big to make a perfect hist plot?
My plot looks like this with separete data and small big.
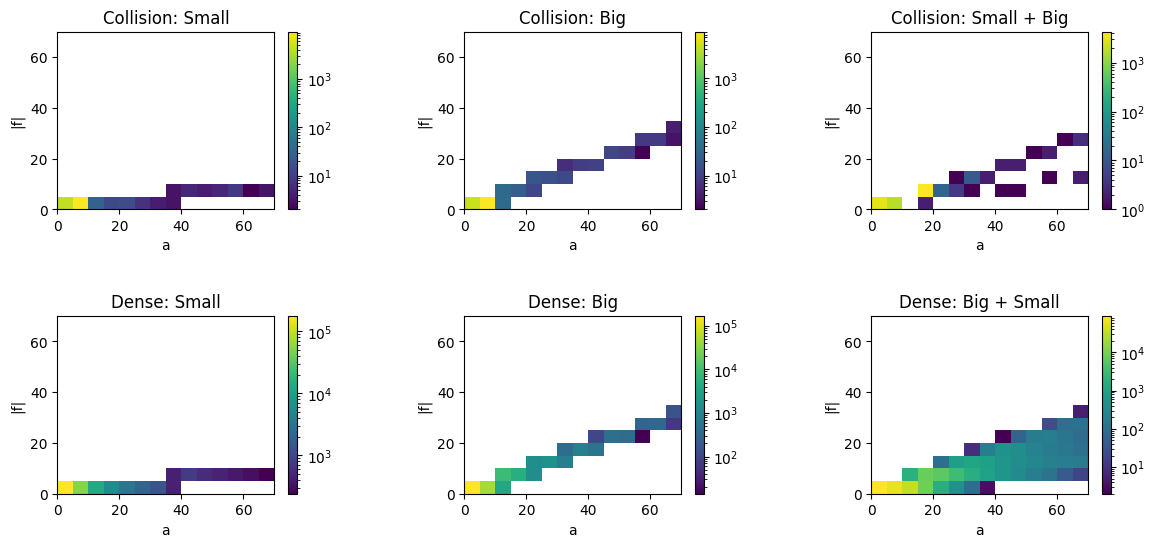
I realized data is missing in collision:Small Big. I was expecting to have plot like Dense:Small Big.
My code is here:
from cProfile import label
from matplotlib.colors import LogNorm
df_collision_big = df_collision[df_collision['type'] == 0]
df_collision_small = df_collision[df_collision['type'] == 1]
df_dense_big = df_dense[df_dense['type'] == 0]
df_dense_small = df_dense[df_dense['type'] == 1]
plt.subplots(figsize=(14, 6))
#make space between subplots
plt.subplots_adjust(wspace=0.5, hspace=0.6)
plt.subplot(231)
plt.hist2d(df_collision_small['a'], df_collision_small['|f|'], bins=np.linspace(0,70,15), norm=LogNorm())
plt.colorbar()
plt.xlabel('a')
plt.ylabel('|f|')
plt.title('Collision: Small')
plt.subplot(232)
plt.hist2d(df_collision_big['a'], df_collision_big['|f|'], bins=np.linspace(0,70,15), norm=LogNorm())
plt.colorbar()
plt.xlabel('a')
plt.ylabel('|f|')
plt.title('Collision: Big')
plt.subplot(233)
plt.hist2d(df_collision_big['a'] df_collision_small['a'], df_collision_big['|f|'] df_collision_small['|f|'], bins=np.linspace(0,70,15), norm=LogNorm())
plt.colorbar()
plt.xlabel('a')
plt.ylabel('|f|')
plt.title('Collision: Small Big')
plt.subplot(234)
plt.hist2d(df_dense_small['a'], df_dense_small['|f|'], bins=np.linspace(0,70,15), norm=LogNorm())
plt.colorbar()
plt.xlabel('a')
plt.ylabel('|f|')
plt.title('Dense: Small')
plt.subplot(235)
plt.hist2d(df_dense_big['a'], df_dense_big['|f|'], bins=np.linspace(0,70,15), norm=LogNorm())
plt.colorbar()
plt.xlabel('a')
plt.ylabel('|f|')
plt.title('Dense: Big')
plt.subplot(236)
plt.hist2d(df_dense_big['a'] df_dense_small['a'], df_dense_big['|f|'] df_dense_small['|f|'], bins=np.linspace(0,70,15), norm=LogNorm())
plt.colorbar()
plt.xlabel('a')
plt.ylabel('|f|')
plt.title('Dense: Big Small')
plt.savefig('hist2d.png', dpi=300)
plt.show()
Printing df_collision['a] gives me this:
175761 9.810009
409899 9.810058
429591 9.810058
358086 9.810009
89079 9.810009
...
243866 9.810058
125778 9.810009
185374 9.810009
496586 9.810058
234942 9.810058
Name: a, Length: 27832, dtype: float64
Most of the values in a are similar.
printing df_collision_big['a'] df_collision_small['a'] gives me this:
0 19.620067
1 19.620067
2 19.620067
3 19.620067
4 19.620067
...
504208 NaN
504209 NaN
504210 NaN
504211 NaN
504212 NaN
Name: a, Length: 18639, dtype: float64
One more thing:
printing len of small and big gives me this:
print(len(df_collision_small['a']))
print(len(df_collision_big['a']))
# Output
13772
14060
Hoping for some suggestions to solve this issue.
Thanks.
CodePudding user response:
You can solve the NaN issue in summation as:
pd.merge(df_collision_small["a"], df_dense_big["a"], how="outer", left_index=True, right_index=True).sum(axis=1)
Explanation
outer join on index results in a dataframe with "a" from both dataframes put together (index/position wise):
df_collision_small = pd.DataFrame({"a" : [1,2]})
>> a
>> 0 1
>> 1 2
df_dense_big = pd.DataFrame({"a" : [10,20,30,40]})
>> a
>> 0 10
>> 1 20
>> 2 30
>> 3 40
pd.merge(df_collision_small, df_dense_big, how="outer", left_index=True, right_index=True)
>> a_x a_y
>> 0 1.0 10
>> 1 2.0 20
>> 2 NaN 30
>> 3 NaN 40
sum(axis=1) ignores NaN while summing values:
pd.merge(df_collision_small["a"], df_dense_big["a"], how="outer", left_index=True, right_index=True).sum(axis=1)
>> 0 11.0
>> 1 22.0
>> 2 30.0
>> 3 40.0
