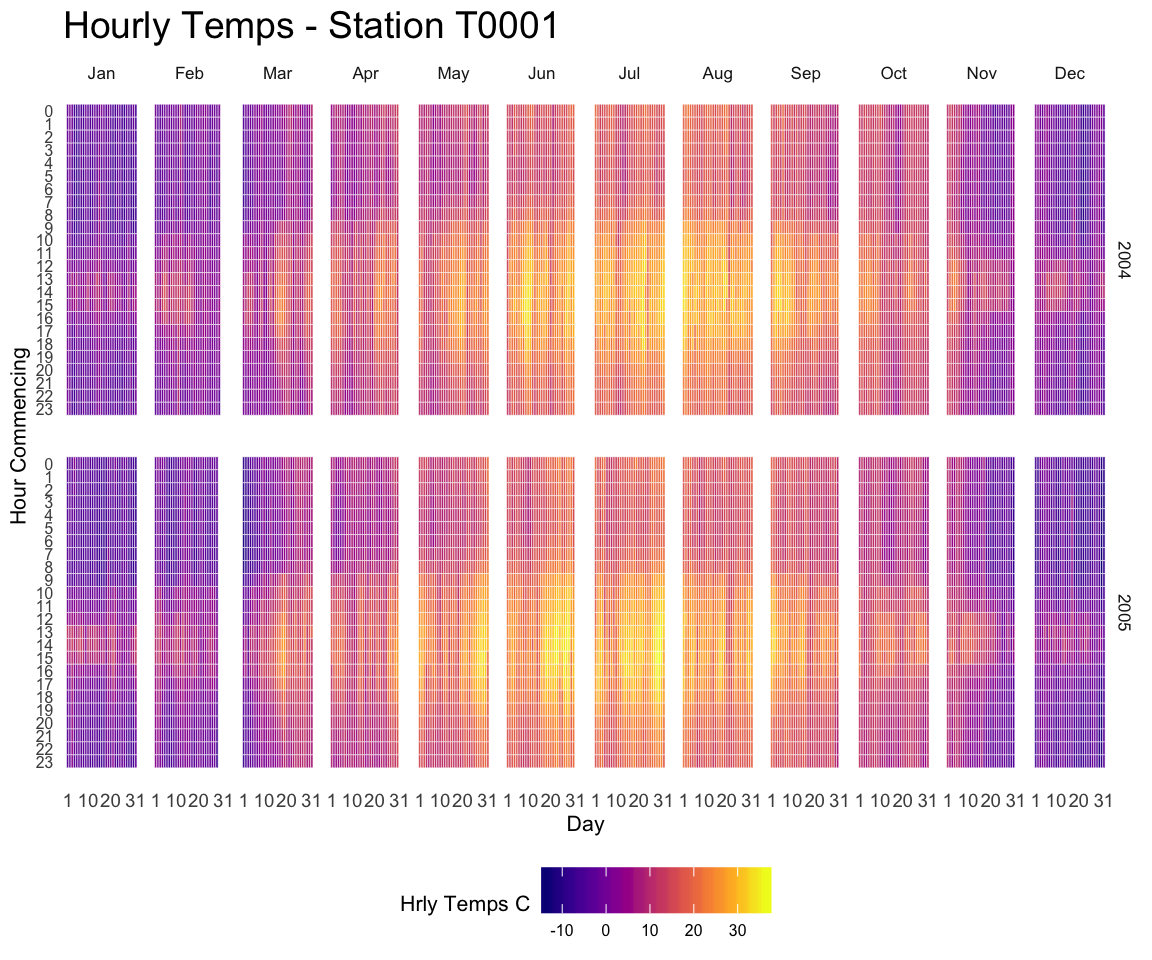
I have a dataset that includes a binary variable (0 vs 1) across time. I want to plot the occurrence of 1 across time. The idea is to see during which period the 1 occurs more often. Here is an example dataset:
set.seed(123)
dd <- data.frame(id = c(rep(1,100), rep(2,80), rep(3,90)),
time = c(seq(from=1,to=100), seq(from=1,to=80), seq(from=1,to=90)),
outcome = c(sample(c(0, 1), 100, replace = TRUE),
sample(c(0, 1), 80, replace = TRUE),
sample(c(0, 1), 90, replace = TRUE)))
My heatmap will have my time variable instead of Day for the x axis and the heat scale will reflect the frequency of 1. For my example dataset, because 1 occurs most often in the second period of time it will be highlighted as the most heated place in the plot.
CodePudding user response:
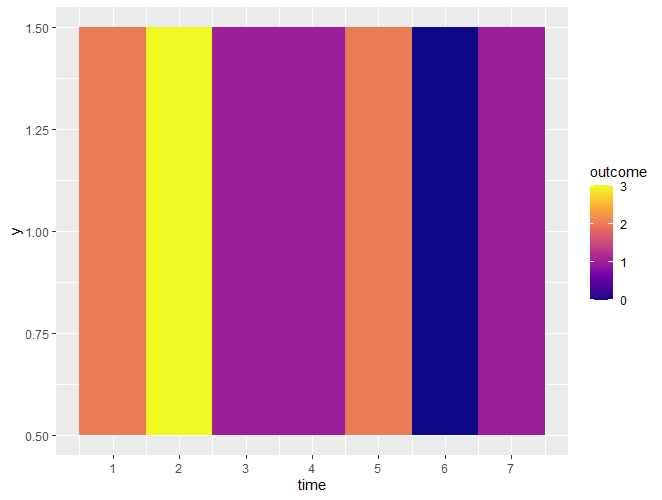
Not sure if I fully understand what you're looking for. Is it something like this:
library(tidyverse)
dd %>%
group_by(time) %>%
summarise(outcome=sum(outcome)) %>%
ggplot(aes(x=time, fill=outcome)) geom_tile(aes(y=1))
scale_x_continuous(breaks = 1:7) scale_fill_viridis_c(option='plasma')
It does highlight the second period as the highest frequency of 1 as required. If you need to split it into months (as per your example image) then use facet_wrap() or facet_grid() with the relevant month variable.
CodePudding user response:
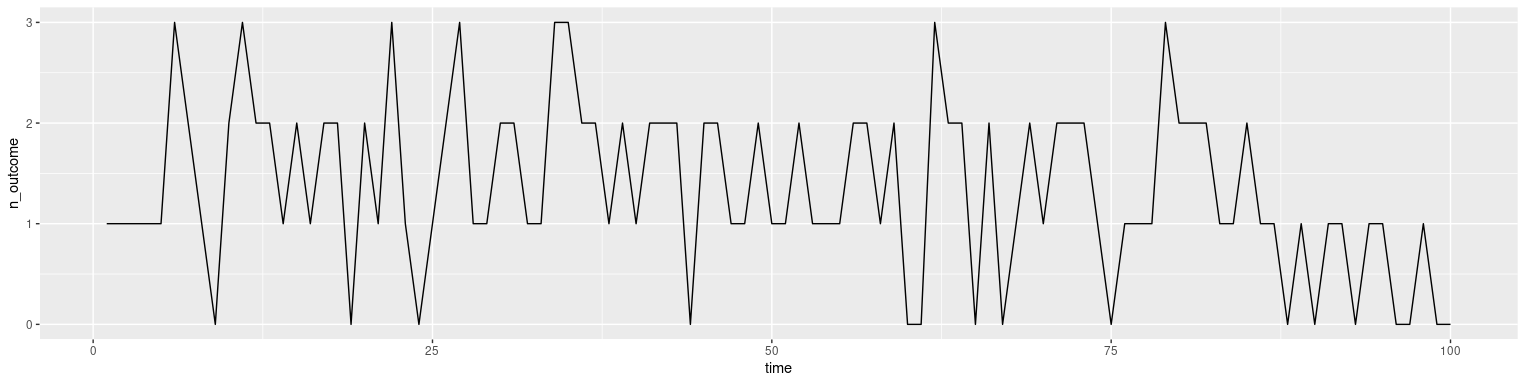
Does something like this work?
dd %>% group_by(time) %>% summarise(n_outcome = sum(outcome)) %>%
ggplot(aes(x = time, y = n_outcome)) geom_line()
You could obviously tweak it to make it prettier