I am really struggling plotting these spatial data points in r. I have tried using ggmap, sf, and sp but I can't get it to cooperate with me. I have a table, tbl, that has the following makeup: tbl
| lat | long | alive| species| where lat and long are the latitude and longitude respectively. "alive" is boolean so is "TRUE" or "FALSE" and "species" is a species code for one of the three species in the data set.
I am trying to get a graph that has points where all of the animals were found, with the color of the point denoting if the animal was found alive or dead and the shape of the point denoting the species. So I could include a key.
I am more familiar with python, and I understand how I could do this in python. But I am really struggling with doing this in r. Could all of these options be passed in the 'aes' parameter? How ould I do that?
Most success doing:
mapplot(longitude=table_1$lon,latitude=table_1$lat,type="p")
mapplot <- get_map(center= c(lon=mean(tbl$lon),lat=mean(tbl$lat)),zoom=2,maptype="satellite",scale=2)
ggmap(mapplot) geom_point(data=tbl, aes(x=lon,y=lat))
CodePudding user response:
Here's a super simple example to get you started, using the sf package for spatial data and ggplot2 for plotting:
require('tidyverse')
require('sf')
# generate some sample data
sampleDF <- data.frame(
lat=c(-17.4, -17.1, -17.8),
lon=c(158.2, 158.9, 157.9),
alive=c(T, F, T),
species=c('sp1', 'sp2', 'sp3')
) %>%
dplyr::mutate(species = as.factor(species)) %>%
st_as_sf(coords=c('lon', 'lat'), crs=4326)
# We converted the species column to a factor
# and converted the dataframe to an sf object,
# specifying the X and Y columns and the
# coordinate reference system (4326 is WGS84)
# Have a look at the sample data
sampleDF
> Simple feature collection with 3 features and 2 fields
> Geometry type: POINT
> Dimension: XY
> Bounding box: xmin: 157.9 ymin: -17.8 xmax: 158.9 ymax: -17.1
> Geodetic CRS: WGS 84
> alive species geometry
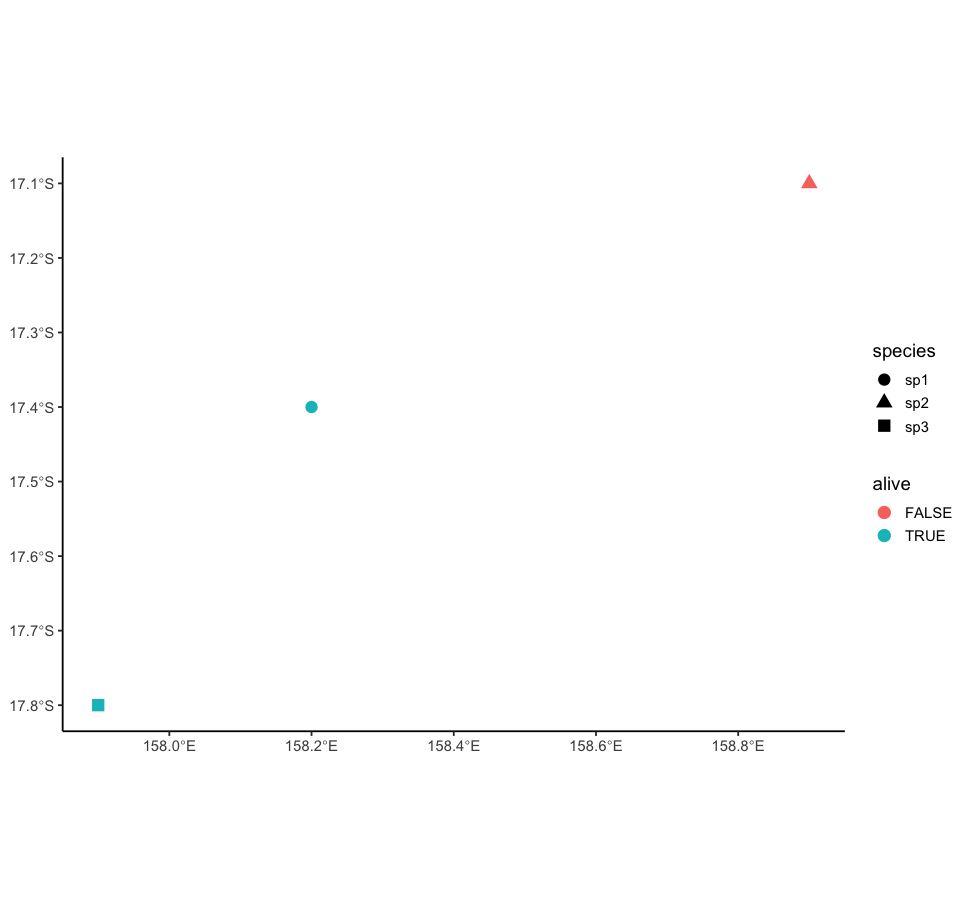
> 1 TRUE sp1 POINT (158.2 -17.4)
> 2 FALSE sp2 POINT (158.9 -17.1)
> 3 TRUE sp3 POINT (157.9 -17.8)
# Now we plot it
# (Note that alive and species are within the aes() function,
# because we want those drawn from the data itself.
# size is outside aes() because we're using a constant of 4.)
ggplot()
geom_sf(data=sampleDF, aes(col=alive, shape=species), size=4)
theme_classic(base_size=14)
Result:
(You can make it prettier by removing the axes, adding a basemap, etc.)
You can also view it interactively with a useful basemap using the mapview package. This will open up a page in your default web browser and let you zoom in/out and change the basemap.
mapview::mapviewOptions(viewer.suppress = TRUE, fgb=FALSE)
mapview::mapview(sampleDF, zcol='alive')
(Note: my random points are in the middle of the Pacific Ocean, so this is not the most useful map.)
CodePudding user response:
Yo, you didn't give us much to go on... so I've taken the liberty of making up some data.
First a table, as you described with lat/lon and alive, species.
Then turn it into an sf object for plotting.
Finally, get some border data to show it on a map.
library(tidyverse)
library(sf)
#> Linking to GEOS 3.8.0, GDAL 3.0.4, PROJ 6.3.1; sf_use_s2() is TRUE
set.seed(3) # for reproducibility
# making up data
lat <- rnorm(10, mean = 36, sd = 4)
long <- rnorm(10, mean = -119, sd = 2)
alive <- sample(c(T, F), 10, replace = T)
species <- sample(c('frog', 'bird', 'rat'), 10, replace = T)
# crate a tibble with the made up data
my_table <- tibble(lat = lat, long = long, alive = alive, species = species)
# turn it into an sf object, for spatial plotting
my_sf <- my_table %>%
st_as_sf(coords = c('long', 'lat')) %>%
st_set_crs(4326) # using 4326 for lat/lon decimal
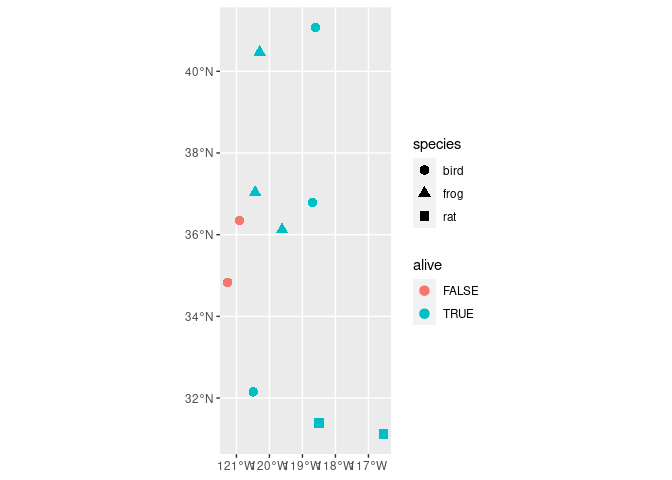
# ggplot2 of the data
ggplot()
geom_sf(data = my_sf, aes(color = alive, shape = species), size = 3)
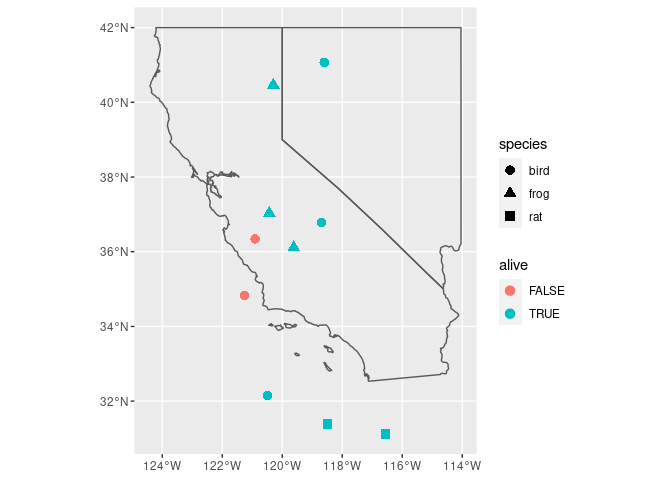
# Getting a little fancier with it by adding the state borders
ca_nv_map <- rnaturalearth::ne_states(country = 'United States of America', returnclass = 'sf') %>%
filter(name %in% c("California", "Nevada"))
ggplot()
geom_sf(data = my_sf, aes(color = alive, shape = species), size = 3)
geom_sf(data = ca_nv_map, fill = NA)
Created on 2022-11-09 by the reprex package (v2.0.1)

