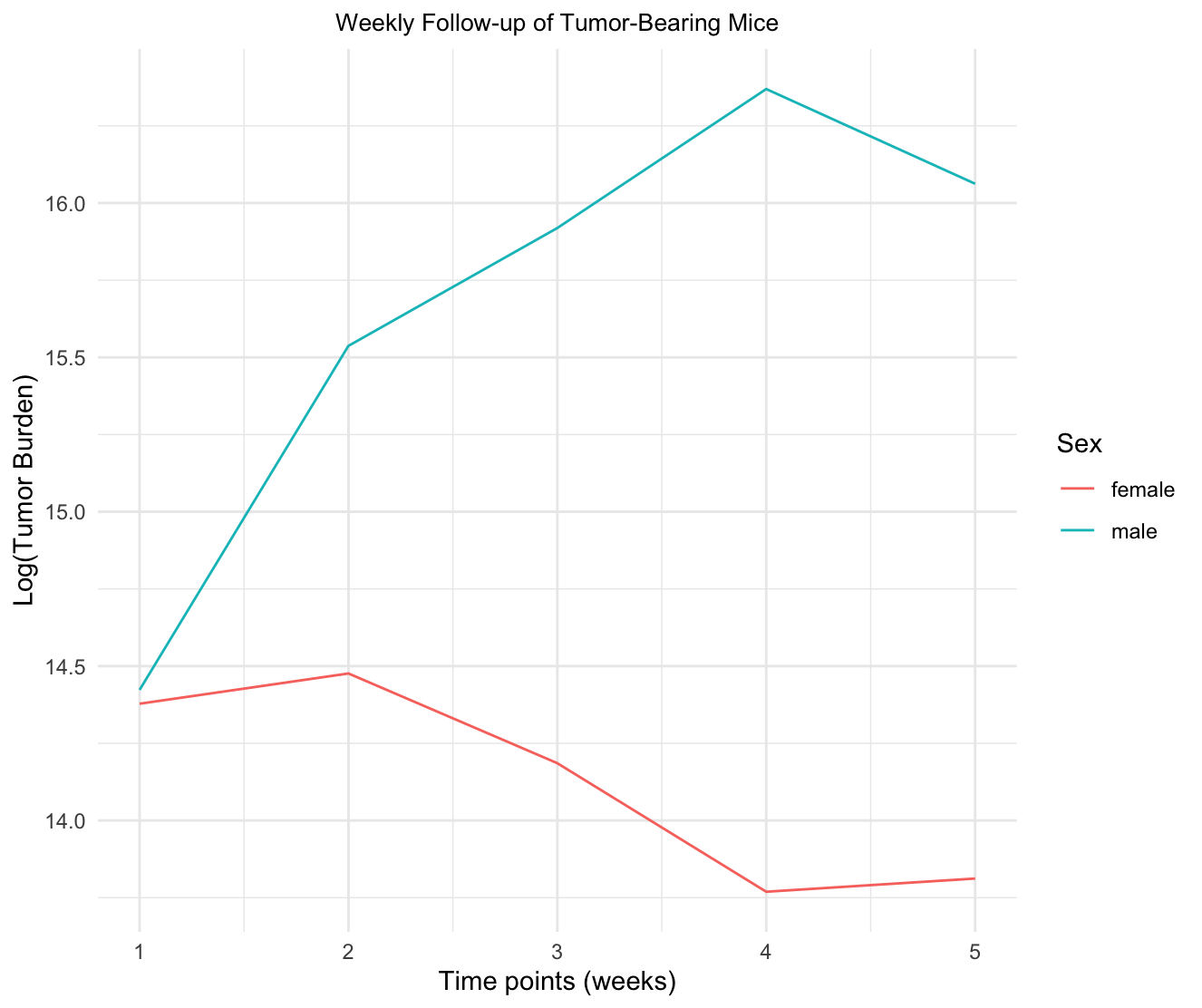
I am looking to display the results of a two-way Anova analysis over several time points. This is preliminary data, and I am interested in getting an understanding of the potential relationship between time and sex on tumor burden.
My data:
ID Sex Tumor.Burden Time.Point
Cage3 female 1270800 1
Cage3 female 1237600 2
Cage3 female 1288760 3
Cage3 female 775220 4
Cage4 female 1768400 1
Cage4 female 1630200 2
Cage4 female 1606900 3
Cage4 female 1134220 4
Cage5 male 1441500 1
Cage5 male 3000750 2
Cage5 male 5930500 3
Cage5 male 6944225 4
Cage6 male 2063640 1
Cage6 male 7067600 2
Cage6 male 10460400 3
Cage6 male 18764800 4
This is the plot I am using. I'd like to point out that this wasn't made with the data I just listed, but rather with similar data. However, I plan on using the same approach here.
ggplot(Data, aes(x = Time.Point, y = Tumor.Burden, color = Sex))
geom_line()
theme_minimal()
labs(title = "Weekly Follow-up of Tumor-Bearing Mice", x = "Time points (weeks)", y="Log(Tumor Burden)")
theme(plot.title = element_text(size = 10, hjust = 0.5))
What is the best approach to add the significance of each time point above to the corresponding time point on the plot? I.E is there a statistically significant difference between males and females at time point 1:5?
Currently, I am following this: https://www.datanovia.com/en/lessons/repeated-measures-anova-in-r/#two-way-repeated-measures-anova. However, I am getting an error at the end and it seems to be related to my ID variable getting flagged as NA when I run
Data %>%
group_by(Time.Point) %>%
anova_test(dv = Tumor.Burden, wid = ID, within = Sex)
Thanks!
CodePudding user response:
To calculate the p values you can use anova_test(Tumor.Burden ~ Sex) and use these output p values in a geom_text to show them in your plot like this:
library(ggplot2)
library(ggpubr)
library(rstatix)
library(dplyr)
p_values <- Data %>%
group_by(Time.Point) %>%
anova_test(Tumor.Burden ~ Sex)
#> Coefficient covariances computed by hccm()
#> Coefficient covariances computed by hccm()
#> Coefficient covariances computed by hccm()
#> Coefficient covariances computed by hccm()
p_values
#> # A tibble: 4 × 8
#> Time.Point Effect DFn DFd F p `p<.05` ges
#> * <int> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
#> 1 1 Sex 1 2 0.342 0.618 "" 0.146
#> 2 2 Sex 1 2 3.11 0.22 "" 0.608
#> 3 3 Sex 1 2 8.83 0.097 "" 0.815
#> 4 4 Sex 1 2 4.05 0.182 "" 0.669
ggplot()
geom_line(Data, mapping = aes(x = Time.Point, y = Tumor.Burden, color = Sex))
geom_text(data = p_values, mapping = aes(x = Time.Point, y = 15000000, label = p), size = 3)
theme_minimal()
labs(title = "Weekly Follow-up of Tumor-Bearing Mice", x = "Time points (weeks)", y="Log(Tumor Burden)")
theme(plot.title = element_text(size = 10, hjust = 0.5))
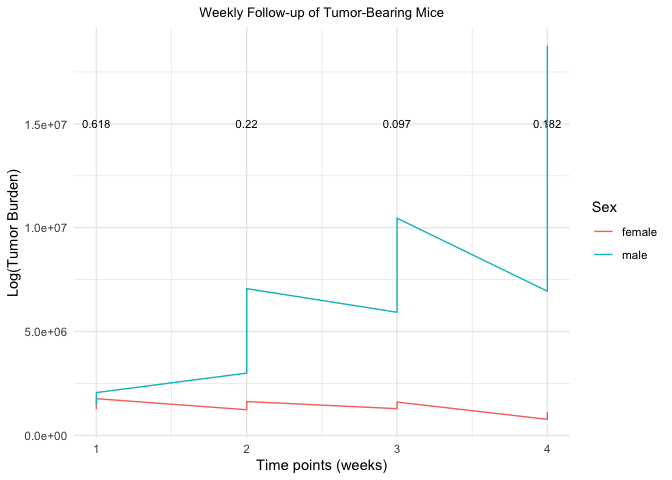
Created on 2022-11-16 with reprex v2.0.2