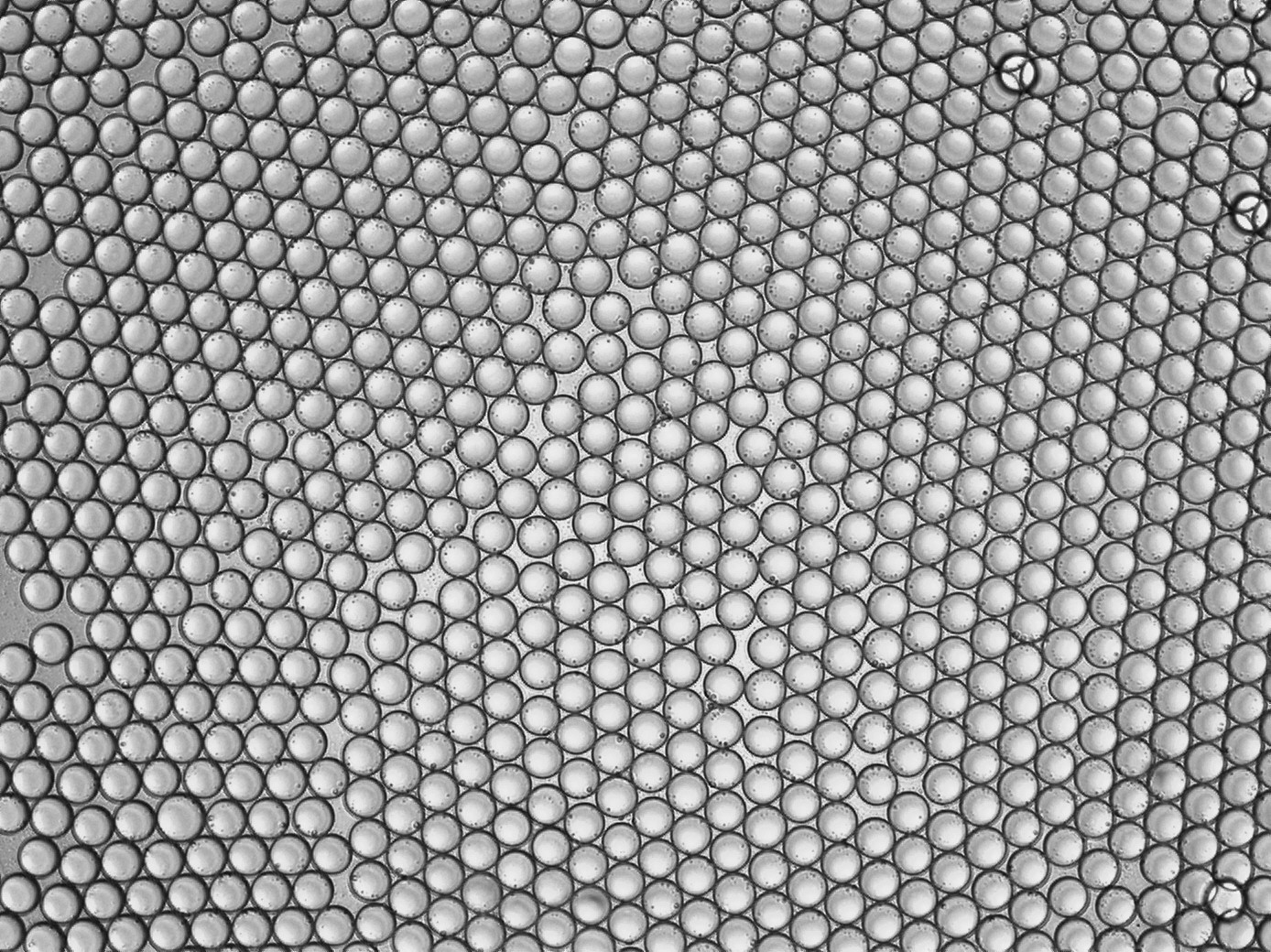
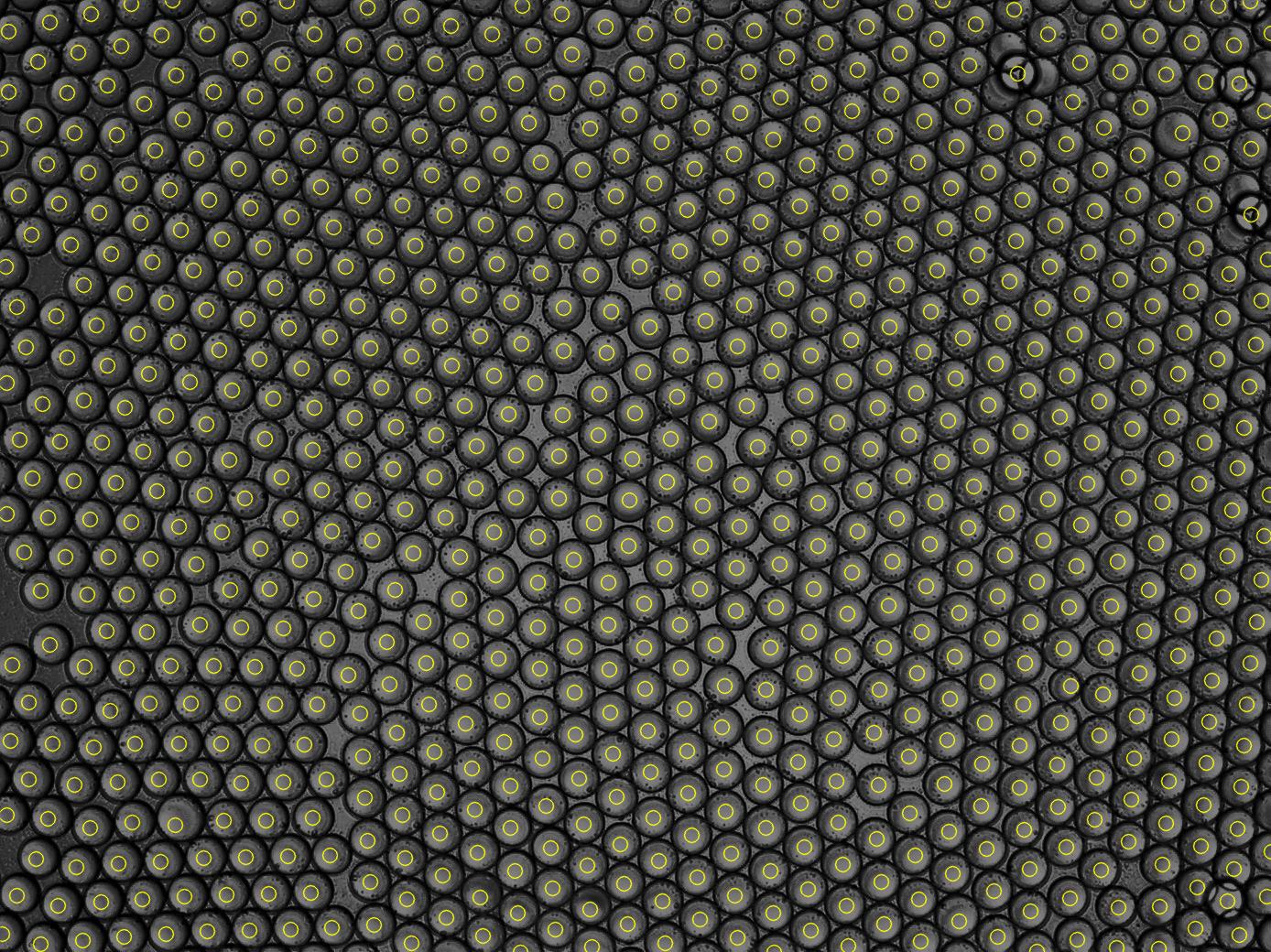
I have image of drops and I want to calculate the number of it.
And Here after threshold application :
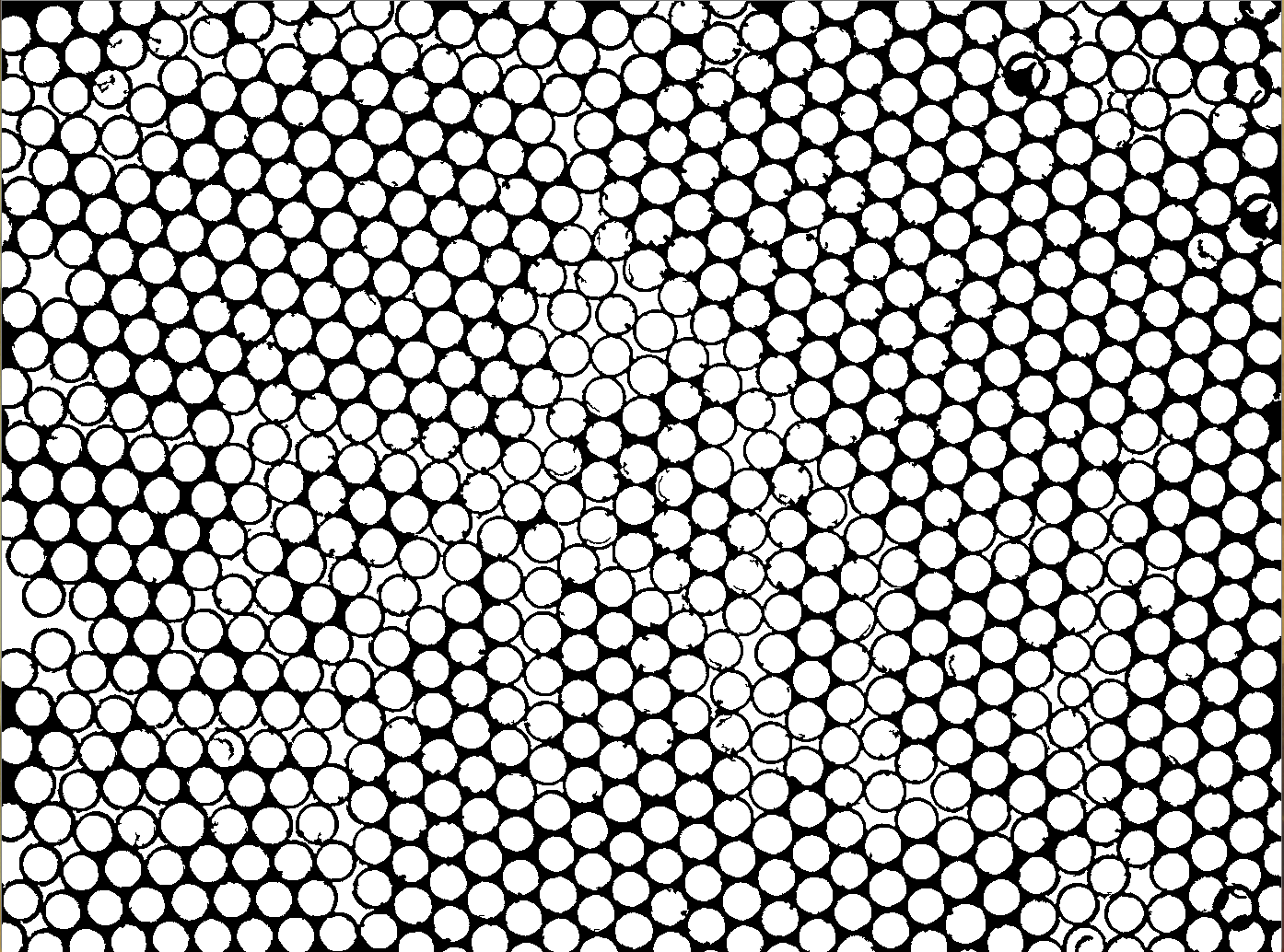
i tried a lot of fonction on OpenCV and it's never right.
Do you have any ideas on how to do ?
Thanks
The best I got, was by using : (img_morph is my binairized image)
rbc_bw = label(img_morph)
rbc_props = regionprops(rbc_bw)
fig, ax = plt.subplots(figsize=(18, 8))
ax.imshow(img_morph)
rbc_count = 0
for i, prop in enumerate(filter(lambda x: x.area > 250, rbc_props)):
y1, x1, y2, x2 = (prop.bbox[0], prop.bbox[1],
prop.bbox[2], prop.bbox[3])
width = x2 - x1
height = y2 - y1
r = plt.Rectangle((x1, y1), width = width, height=height,
color='b', fill=False)
ax.add_patch(r)
rbc_count = 1
print('Red Blood Cell Count:', rbc_count)
plt.show()
And all my circles are detected here but also the gap in between.
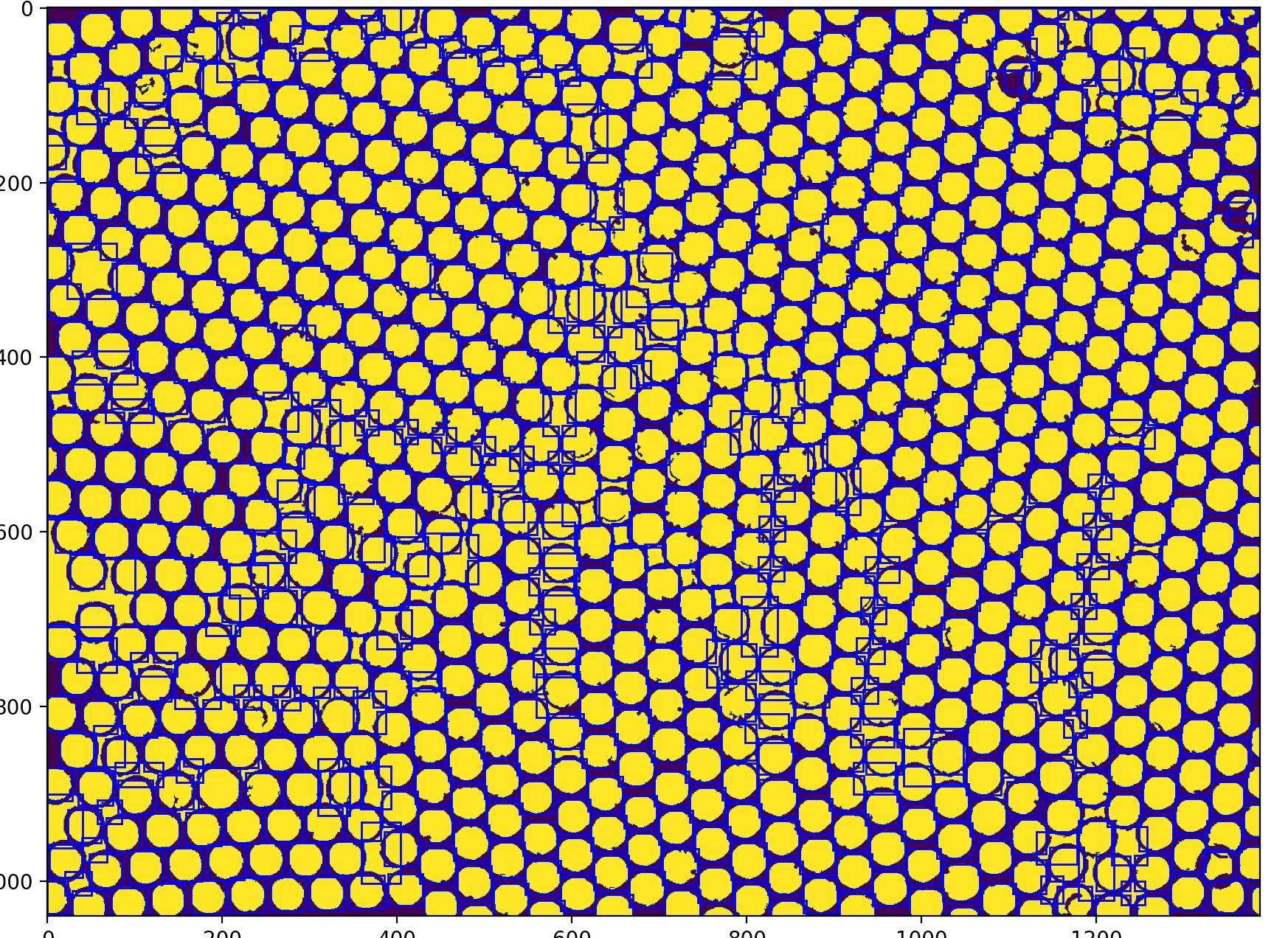
CodePudding user response:
Core idea: matchTemplate.
Approach:
- pick a template manually from the picture
- histogram equalization for badly lit inputs (or always)
matchTemplatewith suitable matching mode- also using
copyMakeBorderto catch instances clipping the border
- also using
- thresholding and non-maximum suppression
I'll skip the boring parts and use the first example input.
Manually picked template:
scores = cv.matchTemplate(haystack, template, cv.TM_CCOEFF_NORMED)
Thresholding and NMS:
levelmask = (scores >= 0.3)
localmax = cv.dilate(scores, None, iterations=26)
localmax = (scores == localmax)
candidates = levelmask & localmax
(nlabels, labels, stats, centroids) = cv.connectedComponentsWithStats(candidates.astype(np.uint8), connectivity=8)
print(nlabels-1, "found") # background counted too
# and then draw a circle for each centroid except label 0
And that finds 766 instances. I see a few false negatives (missed) and saw a false positive too once, but that looks like less than 1%.