I want to create a ggplot for an IBD pedigree analysis (parents/child/siblings) which looks similar to the 23andme produces. I was able to use their IBD phasing algorithm to produce data exemplary for chromosome 20 and some family with ID 745 from the 1000 genome HG38 extendend dataset. The results can be found below as object IBD.The object chrom_sizes gives the length of each chromosome. And the object centromeres gives the start and end position of each chromosome as a reference.
The plot of 23andme looks like the following 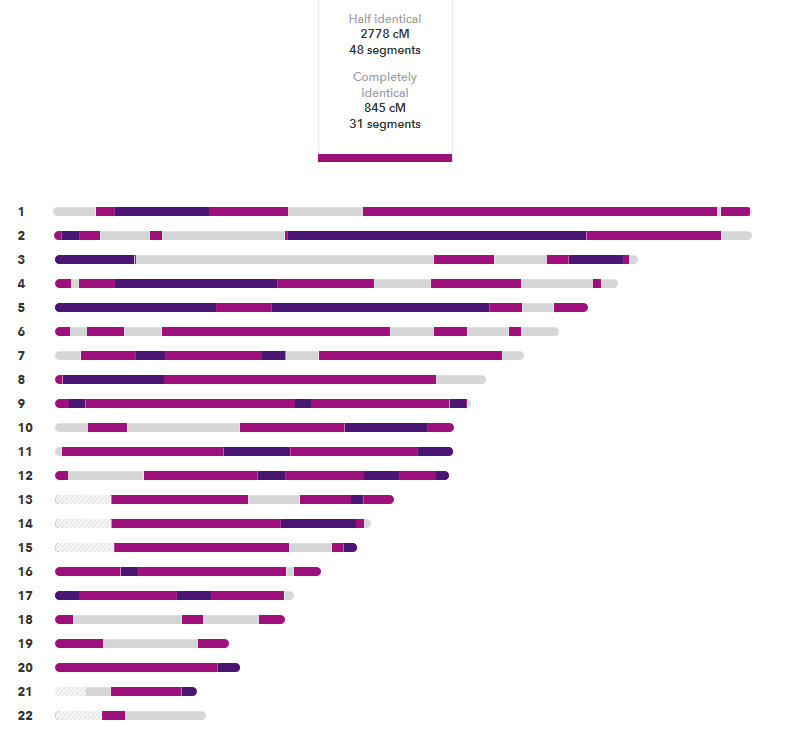
IBD <- tibble(id1 = c(745,745,745,745),
id2 = c(867,1418,1347,1231),
id1_hap =c(1,1,1,0),
id2_hap =c(1,1,0,1),
start=c(1903935, 1917538, 1889345, 1843475),
end=c(1944167, 1943602, 1940182, 1940162 ),
start_cm=c(10.7, 10.8, 10.5, 9.84),
end_cm=c(13.8, 13.8, 13.5, 13.5),
start_bd=c(10676547, 10794199, 10480974, 9836741),
end_bd=c(13839018, 13824389, 13519497, 13518939),
chromosome = c(20, 20, 20, 20)
)
chrom_sizes <- structure(list(chromosome = c("chrM", "chr1", "chr2", "chr3", "chr4",
"chr5", "chr6", "chr7", "chr8", "chr9", "chr10", "chr11", "chr12",
"chr13", "chr14", "chr15", "chr16", "chr17", "chr18", "chr19",
"chr20", "chr21", "chr22", "chrX", "chrY"),
size = c(248956422L, 242193529L, 198295559L, 190214555L, 181538259L,
170805979L, 159345973L, 145138636L, 138394717L, 133797422L,
135086622L, 133275309L, 114364328L, 107043718L, 101991189L,
90338345L, 83257441L, 80373285L, 58617616L, 64444167L, 46709983L,
50818468L, 156040895L, 57227415L, 16569L)),
.Names = c("chromosome","size"),
class = "data.frame", row.names = c(NA, -25L))
# hg19 centromere locations
centromeres <- structure(list(chromosome = c("chr1", "chr2", "chr3", "chr4",
"chr5", "chr6", "chr7", "chr8", "chr9", "chrX", "chrY", "chr10",
"chr11", "chr12", "chr13", "chr14", "chr15", "chr16", "chr17",
"chr18", "chr19", "chr20", "chr21", "chr22"),
start = c(121535434L, 92326171L, 90504854L, 49660117L, 46405641L, 58830166L,
58054331L, 43838887L, 47367679L, 58632012L, 10104553L, 39254935L, 51644205L,
34856694L, 16000000L, 16000000L, 17000000L, 35335801L, 22263006L,
15460898L, 24681782L, 26369569L, 11288129L, 13000000L),
end = c(124535434L,95326171L, 93504854L, 52660117L, 49405641L, 61830166L,
61054331L, 46838887L, 50367679L, 61632012L, 13104553L, 42254935L,
54644205L, 37856694L, 19000000L, 19000000L, 20000000L, 38335801L,
25263006L, 18460898L, 27681782L, 29369569L, 14288129L, 16000000L)),
.Names = c("chromosome","start", "end"),
class = "data.frame", row.names = c(NA, -24L))
I'm aware that my data only contains overlap for the chromosome 20 but I would love to get an outline for all other chromosomes aswell despite the bars would be appear without filled in values.
Q: How can one create IBD anylsis plots in the style 23andme produces them?
CodePudding user response:
Like Konrad mentions, it's mostly geom_segments. To get you started, here is a way that it could work, though I was unsure which part of your data to use as colour.
# Data construction same as in question, omitted for brevity
ggplot()
geom_segment(
data = chrom_sizes,
aes(x = 0, xend = size, y = chromosome, yend = chromosome),
linewidth = 4, lineend = "round", colour = "grey75"
)
geom_segment(
data = centromeres,
aes(x = start, xend = end, y = chromosome, yend = chromosome),
linewidth = 4, colour = "grey85"
)
geom_segment(
data = IBD,
aes(
x = start_bd, xend = end_bd,
y = paste0("chr", chromosome), yend = paste0("chr", chromosome),
colour = id1_hap == id2_hap
),
linewidth = 4
)
scale_y_discrete(
limits = unique(chrom_sizes$chromosome)
)
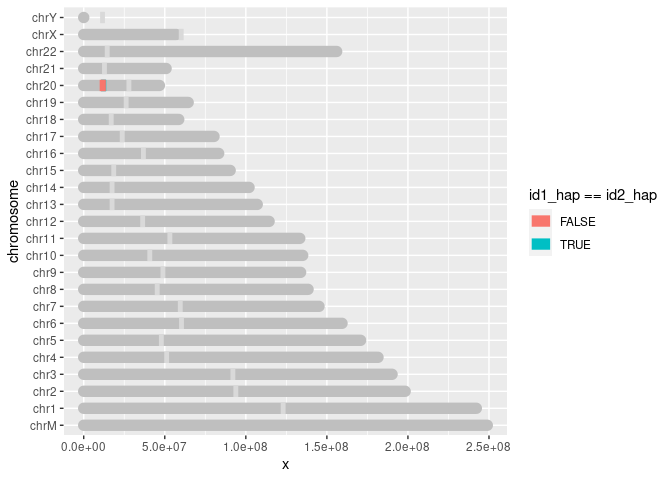
You can also use limits = rev(unique(chrom_sizes$chromosome)) to change the sorting of chromosomes.
As an aside, I think you'd want to double check your data because it seems implausible to me that any organism has such a huge mitochondrial genome. Moreover, according to your data, the centromere of the Y chromosome falls outside the bounds of the chromosome, which seems unlikely to me.
