As a follow up on my experiments, I need to increase the reagents I want to test. I was able to create a for loop which takes the concentration and amounts of a excel file, and creates pivot tables/data frames which show me the concentration as Index/Header and the amounts to pipet in the cells. However, I struggle at saving them into a single worksheet since they are multiple dataframes.
My current approach results in an TypeError: unhashable type: 'DataFrame'
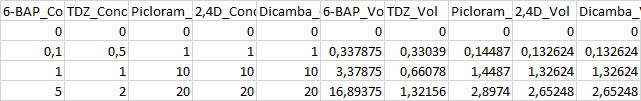
Here is the starting data:
| 6-BAP_Conc | TDZ_Conc | Picloram_Conc | 2,4D_Conc | Dicamba_Conc | 6-BAP_Vol | TDZ_Vol Picloram_Vol | 2,4D_Vol | Dicamba_Vol |
|---|---|---|---|---|---|---|---|---|
| 0.0 | 0.0 | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 0.1 | 0.5 | 1 | 1 | 1 | 0.337875 | 0.33039 | 0.14487 | 0.132624 |
| 1.0 | 1.0 | 10 | 10 | 10 | 3.37875 | 0.6607799999999999 | 1.4487 | 1.32624 |
| 5.0 | 2.0 | 20 | 20 | 20 | 16.89375 | 1.32156 | 2.8974 | 2.65248 |
So here is my approach:
import numpy as np
import pandas as pd
from itertools import product
df = pd.read_excel("BlueTest.xlsx")
with pd.ExcelWriter('test3.xlsx', engine='xlsxwriter') as writer:
workbook = writer.book
worksheet = workbook.add_worksheet('Result')
writer.sheets['Result'] = worksheet
COLUMN = 0
row = 0
for x in ["6-BAP", "TDZ"]:
for y in ["Picloram","2,4D","Dicamba"]:
df1 = df.loc[:,df.columns.str.startswith(x)]
df2 = df.loc[:,df.columns.str.startswith(y)]
my_product = list(product(df1[f"{x}_Vol"], df2[f"{y}_Vol"]))
my_product_str = [str(a) "uL" for a in my_product]
my_product_str_np = np.array(my_product_str)
my_product_str_np = my_product_str_np.reshape(len(df1), len(df2))
dfn = pd.DataFrame(my_product_str_np, index=df1[f"{x}_Conc.1"], columns=df2[f"{y}_Conc"])
worksheet.write_string(row,COLUMN,dfn)
row = 1
dfn.to_excel(writer, sheet_name="Results", startrow= row, startcol=COLUMN)
row = dfn.shape[0] 2
I would really appreciate some help with this, since im struggeling hard with these kind of things and python.
CodePudding user response:
This works for me
import numpy as np
import pandas as pd
from itertools import product
#reading the table you pasted
df = pd.read_clipboard()
#the function that will append new data or create an empty dataframe
def append_df(df_data, df_result):
if df_data.empty:
df_data = df_result
else:
df_data = df_data.append(other=df_result, ignore_index=True, sort=False)
return df_data
#seems you have and error in the table so here I am sorting it out (error when comparing table to the picture(TDZ_Vol Picloram_Vol are in one column and not separate))
df=df.rename(columns={"TDZ_Vol Picloram_Vol":"TDZ_Vol"})
df = df.assign(Picloram_Vol=[0,0.14487,0.14487,2.8974])
df=df.rename(columns={"col_name":"Picloram_Vol"})
df_data=pd.DataFrame()
for x in ["6-BAP", "TDZ"]:
for y in ["Picloram","2,4D","Dicamba"]:
df1 = df.loc[:,df.columns.str.startswith(x)]
df2 = df.loc[:,df.columns.str.startswith(y)]
my_product = list(product(df1[f"{x}_Vol"], df2[f"{y}_Vol"]))
my_product_str = [str(a) "uL" for a in my_product]
my_product_str_np = np.array(my_product_str)
my_product_str_np = my_product_str_np.reshape(len(df1), len(df2))
dfn = pd.DataFrame(my_product_str_np, index=df1[f"{x}_Conc"], columns=df2[f"{y}_Conc"])
#reseting index as concentrations can be different
dfn=dfn.reset_index()
#reating a row with your column names (as concetrations can be different )
dfn=pd.DataFrame(np.vstack([dfn.columns, dfn]))
#using the function top append data
df_data = append_df(df_data, dfn)