I have multiple files. Each corresponding to the data of a city having an ID and "VOLUME" column. I am trying to create dataframes from csv files and after treatments i would like to merge these dataframes by "ID" with all "VOLUME" columns like this : VOLUME_{CITY} with {CITY} corresponding to the name of the city.
So far i did this :
data_dir_file = 'Data'
cities = ['NY','Chicago','Boston','Toronto']
dfs = []
for city in cities:
file_name = f'Data_2010_{city}.csv'
df = pd.read_csv(f'{data_dir_file}\{file_name}',sep=';')
df = df[['ID','VOLUME']].drop_duplicates()
dfs.append(df)
df_concat = pd.concat(dfs)
print(df_concat)
So this concatenates each dataframe but how can i merge them by "ID" instead of doing a concatenation ?
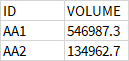
Each dataframe looks like this
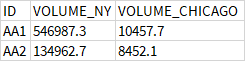
What i want is something like this :
CodePudding user response:
You can use pd.merge to merge your data by the ID (also change the name of column "VOLUME", otherwise they'll be renamed as VOLUME_x, VOLUME_y losing information about the city)
data_dir_file = 'Data'
cities = ['NY','Chicago','Boston','Toronto']
dfs = pd.DataFrame()
for city in cities:
file_name = f'Data_2010_{city}.csv'
df = pd.read_csv(f'{data_dir_file}\{file_name}',sep=';')
df = df[['ID','VOLUME']].drop_duplicates()
df.columns = ['ID','VOLUME_' city]
if len(dfs) == 0:
dfs = df
else:
dfs = dfs.merge(df, on= 'ID', outer=True)
CodePudding user response:
If you remove duplicates on 'ID', you can define ID column as your index then concat all dataframes together.
import pathlib
data_dir_file = pathlib.Path('Data')
cities = ['NY', 'Chicago', 'Boston', 'Toronto']
dfs = []
for city in cities:
file_name = f'Data_2010_{city}.csv'
df = pd.read_csv(data_dir_file / file_name, sep=';')
df = (df[['ID', 'VOLUME']].drop_duplicates('ID').set_index('ID')
.squeeze().rename(f'VOLUME_{city}'))
dfs.append(df)
# Consider ID is unique after drop_duplicates else use merge reduction
df_concat = pd.concat(dfs, axis=1)