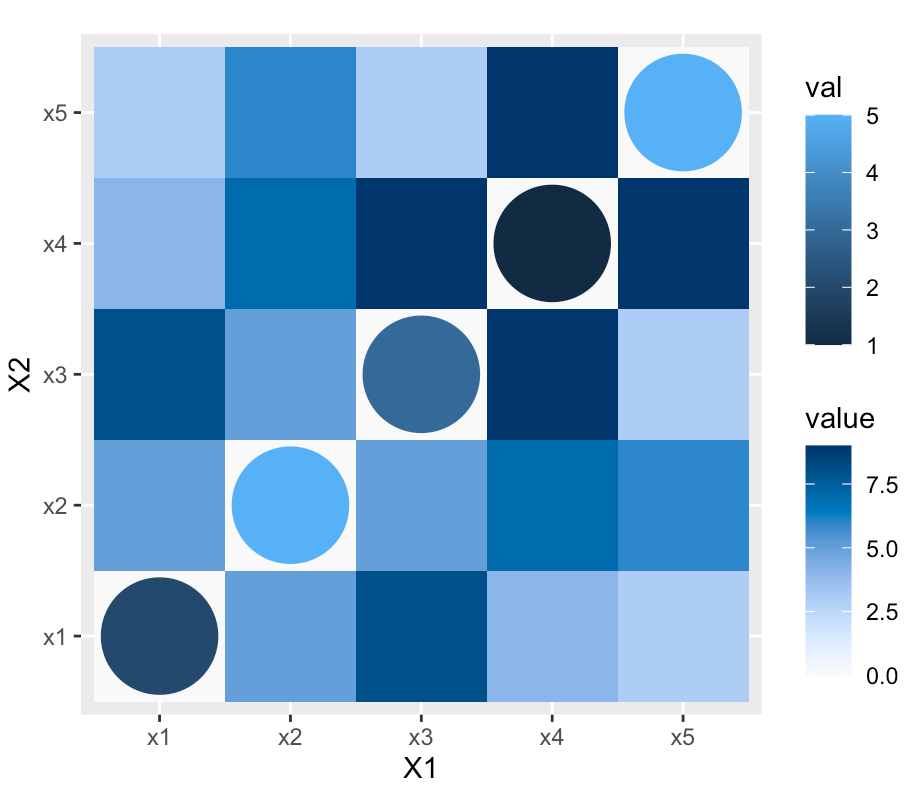
Basically, I have a heatmap that contains some points. What Im trying to do is automatically rescale the size of the points in a sensible way for different sized heatmaps. For example, if I have a heatmap that looks like so:
library(reshape)
library(ggplot2)
library(ggnewscale)
# Create matrix
set.seed(1701)
a <- sample(1:10,100, replace=TRUE)
s <- matrix(a, nrow = 5, ncol=5)
s[upper.tri(s)] = t(s)[upper.tri(s)]
rownames(s) <- colnames(s) <- paste0("x", 1:5)
diag(s) <- 0
sDf <- melt(s)
# create diagonal values
diagDf <- data.frame(
var1 = c(paste0("x", 1:5)),
var2 = c(paste0("x", 1:5)),
val = c(2,5,3,1,5)
)
# make plot
ggplot(sDf, aes(X1,X2))
geom_tile(aes(fill = value))
scale_fill_gradientn(colors = rev(colorspace::sequential_hcl(palette = "Blues 3", n = 100)))
new_scale_fill()
geom_point(data = diagDf, aes(var1, var2, col = val), size = 20)
theme(aspect.ratio = 1)
So in the image above, the diagonal contains geom_points and their size is manually set to size = 20.... This works for this example, but the issue is:
If the heatmap dimensions were changed to say 20x20, then having the size hardcoded to equal 20 won't work due to overlapping & the points being too big etc.
So what Im trying to do is come up with a method that will automatically resize the points to effectively fill square they are contained in without overlapping, being too big or too small.
Any suggestions as to how I could do this?
CodePudding user response:
I would do something like this:
library(reshape)
library(ggplot2)
library(ggnewscale)
n <- 5
# Create matrix
set.seed(1701)
a <- sample(1:10,100, replace=TRUE)
s <- matrix(a, nrow = n, ncol=n)
s[upper.tri(s)] = t(s)[upper.tri(s)]
rownames(s) <- colnames(s) <- paste0("x", 1:n)
diag(s) <- 0
sDf <- melt(s)
# create diagonal values
diagDf <- data.frame(
var1 = c(paste0("x", 1:n)),
var2 = c(paste0("x", 1:n)),
val = sample(1:5,n,replace = T)
)
# make plot
ggplot(sDf, aes(X1,X2))
geom_tile(aes(fill = value))
scale_fill_gradientn(colors = rev(colorspace::sequential_hcl(palette = "Blues 3", n = 100)))
new_scale_fill()
geom_point(data = diagDf, aes(var1, var2, col = val), size = 1/sqrt(nrow(sDf))*80)
theme(aspect.ratio = 1)
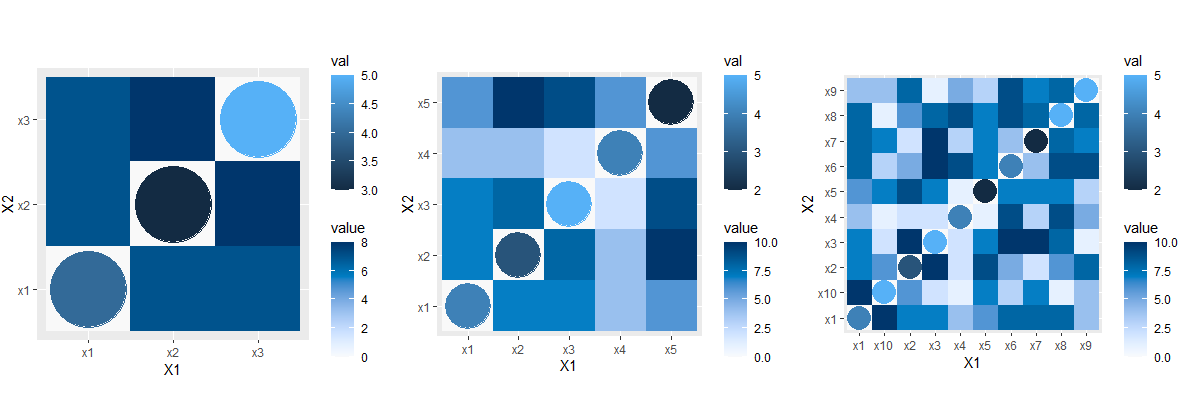
here the size of the points depends on the dimension of the matrix.
an example of the output with a 3x3, 5x5, and 10x10 matrix
CodePudding user response:
You can modify diagDf to contain the co-ordinates of the circles you want to plot using some basic trigonometry, then plot them as filled polygons. This ensures they will always scale exactly with your plot.
library(dplyr)
diagDf <- diagDf %>%
mutate(var1 = as.numeric(as.factor(var1)),
var2 = as.numeric(as.factor(var2))) %>%
split.data.frame(diagDf$var1) %>%
lapply(function(x) {
deg <- seq(0, 2 * pi, length = 100)
var1 <- cos(deg)/2.2
var2 <- sin(deg)/2.2
val <- rep(x$val, 100)
data.frame(var1 = var1 x$var1, var2 = var2 x$var2, val = val)}) %>%
{do.call(rbind, .)}
Now with slightly modified plot code, we get:
ggplot(sDf, aes(X1,X2))
geom_tile(aes(fill = value))
scale_fill_gradientn(colors=rev(colorspace::sequential_hcl(palette = "Blues 3", n=100)))
new_scale_fill()
geom_polygon(data = diagDf, aes(var1, var2, fill = val, group = val))
theme(aspect.ratio = 1)
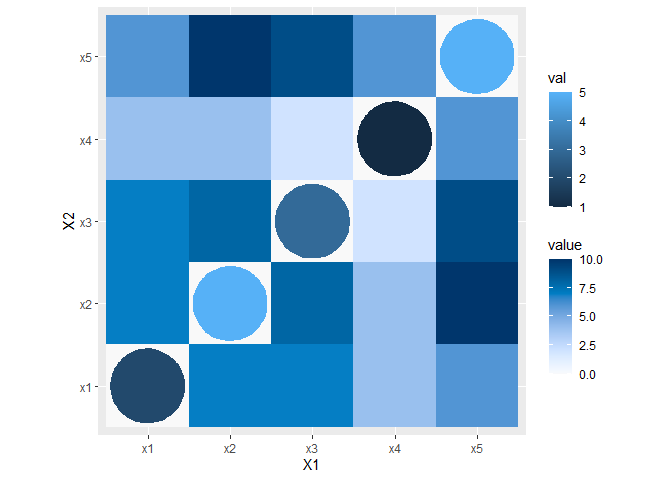
Created on 2021-09-27 by the reprex package (v2.0.0)