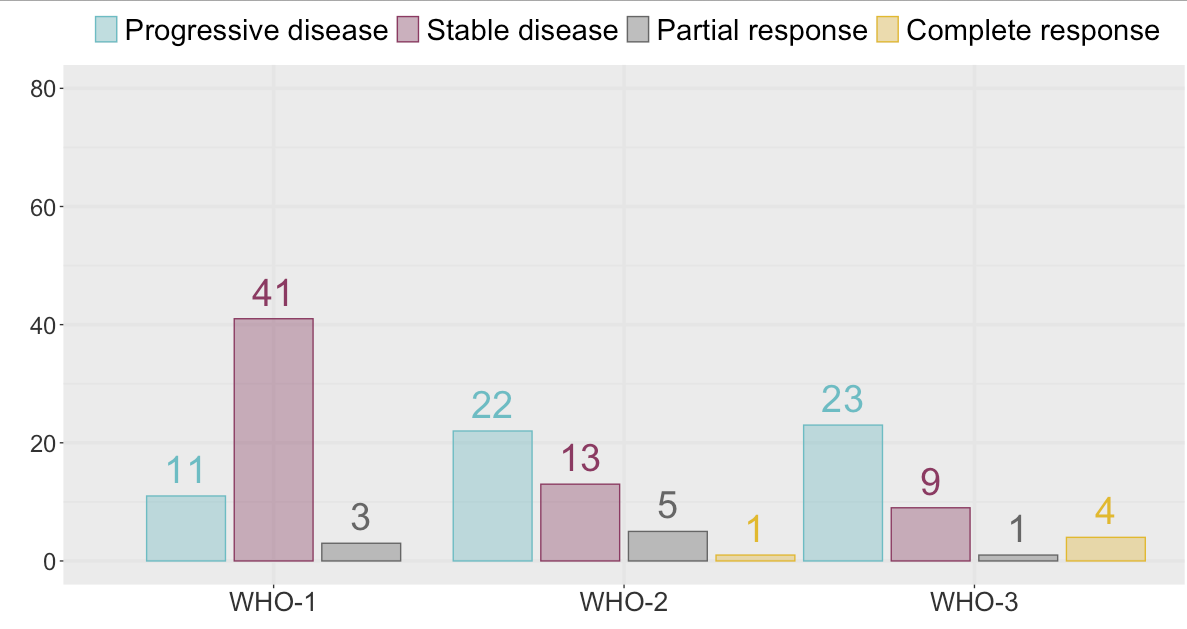
Lets say I have
As you can see, there are 0 Complete Response in WHO-1, but there are cases in the other categories. So, it looks graphically off that there are three geom_col-bars in the WHO-1-category and four geom_col-bars in the other two.
How can I add a fourth bar to WHO-1 indicating the space corresponding to Complete Response?
Something like
Script
pp %>%
as_tibble() %>%
mutate(nyWHO = as.factor(WHO),
best.resp = as.factor(case_when(best_rad == "CR" ~ 4,
best_rad == "PR" ~ 3,
best_rad == "SD" ~ 2,
best_rad == "PD" ~ 1))) %>%
count(nyWHO, best.resp) %>%
ggplot(aes(nyWHO, n, color = best.resp, fill= best.resp))
scale_fill_manual(values = alpha(c("#6DBCC3", "#8B3A62", "grey40", "#E1B930"), 0.4),
name="",
labels = c("Progressive disease", "Stable disease", "Partial response", "Complete response"))
scale_colour_manual(values = cols,
name="",
labels = c("Progressive disease", "Stable disease", "Partial response", "Complete response"))
geom_col(width=1, position = position_dodge2(width = 1, preserve = "single"))
geom_text(aes(label = n), position = position_dodge2(width = 1, preserve = "single"),
vjust=-0.5, size = 10, show.legend = F)
scale_x_discrete(name = "", labels = c("WHO-1", "WHO-2", "WHO-3"))
scale_y_continuous(name="", breaks = seq(0, 80, by = 20))
coord_cartesian(ylim=c(0, 80))
theme(axis.title.x = element_text(size = 22),
axis.title.y = element_text(size = 22),
axis.text.x = ggtext::element_markdown(color = "grey20", size = 20),
axis.text.y = element_text(color = "grey20", size = 18),
panel.grid.major = element_line(colour = "gray90", size = 1.2),
panel.grid.minor = element_line(colour = "gray90", size = 0.6),
legend.text = element_text(size = 22),
legend.position = "top")
Data
pp <- structure(list(WHO = structure(c(3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 1L, 3L, 1L, 3L, 3L, 2L, 3L, 1L, 1L, 2L, 1L, 1L, 3L, 2L, 1L, 1L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 2L, 3L, 2L, 1L, 2L, 3L, 2L, 2L, 2L, 1L, 3L, 3L, 3L, 2L, 3L, 2L, 3L, 1L, 1L, 1L, 3L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 3L, 2L, 3L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 3L, 1L, 1L, 3L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 3L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 3L, 3L, 3L, 3L, 3L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 3L, 2L, 2L, 2L, 3L, 2L, 3L, 3L), .Label = c("1", "2", "3"), class = "factor"), best_rad = c("SD", "SD", "CR", "CR", "SD", "SD", "SD", "SD", "PR", "PR", "PR", "PR", "CR", "CR", "PR", "PR", "PR", "SD", "PD", "PR", "PD", "SD", "SD", "PD", "PD", "PD", "PD", "SD", "PD", "PR", "PD", "PD", "PD", "PD", "SD", "PD", "SD", "SD", "PD", "PD", "PD", "PD", "PD", "PD", "PD", "PD", "PD", "PD", "PD", "PD", "SD", "PD", "PD", "PD", "PD", "SD", "PD", "PD", "SD", "SD", "SD", "PD", "SD", "PD", "SD", "SD", "PD", "PD", "SD", "PD", "PD", "PD", "SD", "SD", "SD", "SD", "PD", "SD", "PD", "SD", "PD", "SD", "PD", "SD", "PD", "SD", "SD", "PD", "SD", "SD", "SD", "PD", "SD", "SD", "SD", "PD", "PD", "PD", "SD", "SD", "SD", "CR", "SD", "SD", "SD", "PD", "PD", "PD", "SD", "SD", "SD", "SD", "SD", "SD", "SD", "SD", "SD", "SD", "SD", "SD", "SD", "SD", "SD", "SD", "PD", "PD", "SD", "SD", "PD", "PD", "PD", "SD", "PD")), row.names = c(NA, -133L), class = "data.frame")
CodePudding user response:
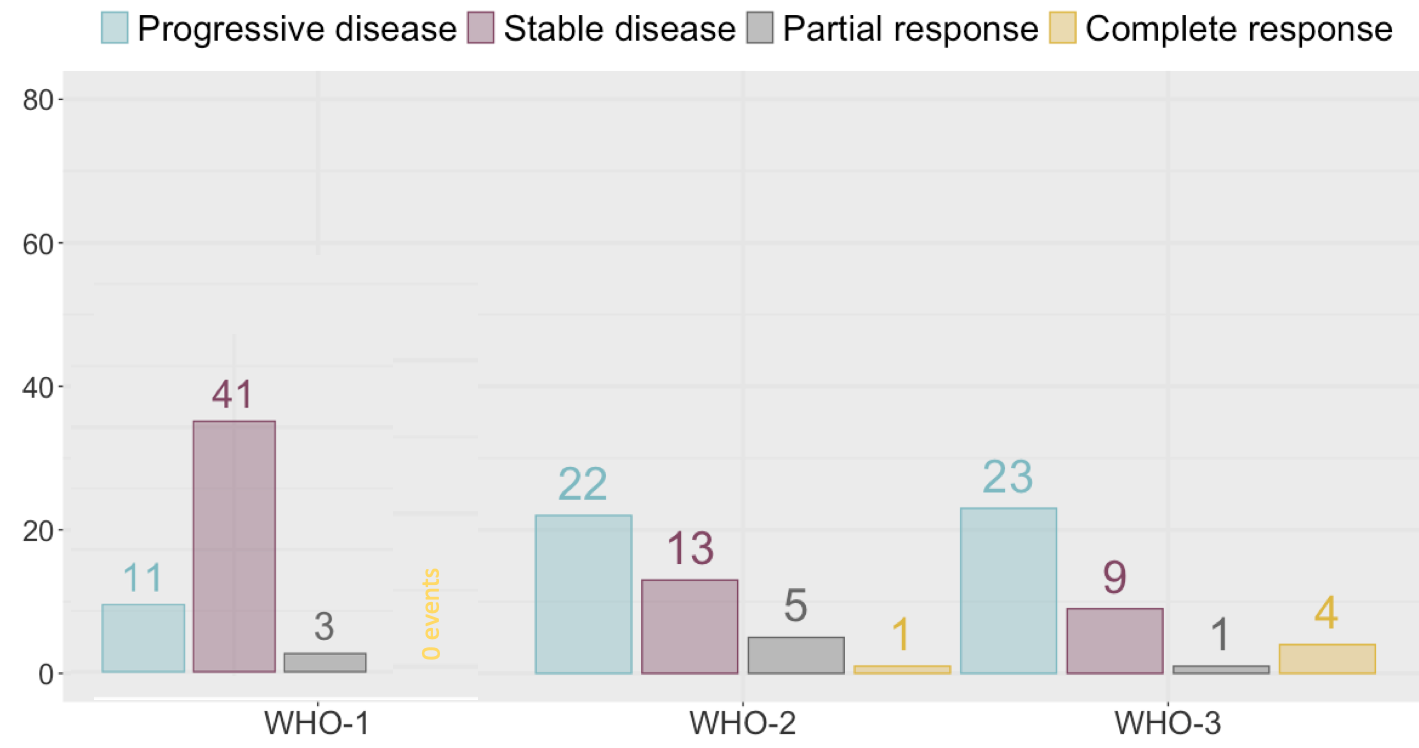
Use count(nyWHO, best.resp, .drop = FALSE)
d <- pp %>%
as_tibble() %>%
mutate(nyWHO = as.factor(WHO),
best.resp = as.factor(case_when(best_rad == "CR" ~ 4,
best_rad == "PR" ~ 3,
best_rad == "SD" ~ 2,
best_rad == "PD" ~ 1))) %>%
count(nyWHO, best.resp, .drop = FALSE)
d
# A tibble: 12 x 3
nyWHO best.resp n
<fct> <fct> <int>
1 1 1 11
2 1 2 41
3 1 3 3
4 1 4 0
5 2 1 22
6 2 2 13
7 2 3 5
8 2 4 1
9 3 1 23
10 3 2 9
11 3 3 1
12 3 4 4
ggplot(...)
CodePudding user response:
Using .drop = FALSE is the solution, but if not possible say, the raw data is unavailable, then a solution is to expand the data for all the levels by using a join:
...
data <- pp %>%
as_tibble() %>%
mutate(nyWHO = as.factor(WHO),
best.resp = as.factor(case_when(best_rad == "CR" ~ 4,
best_rad == "PR" ~ 3,
best_rad == "SD" ~ 2,
best_rad == "PD" ~ 1))) %>%
count(nyWHO, best.resp)
data %>%
full_join(expand(data, nyWHO, best.resp), by = c("nyWHO", "best.resp")) %>%
replace_na(list(n = 0)) %>%
ggplot(aes(nyWHO, n, color = best.resp, fill= best.resp))
... # rest of ggplot statement