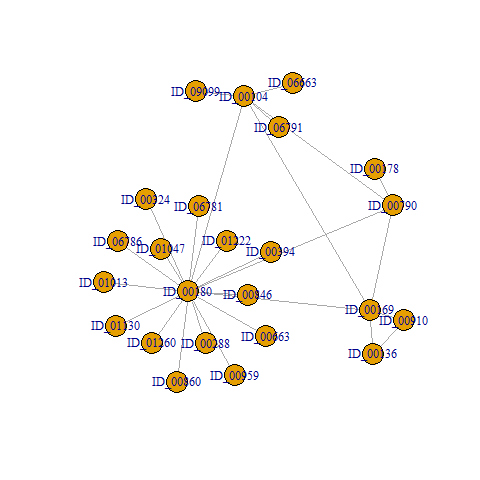
I have a graph that contains 4 components of different cluster sizes.
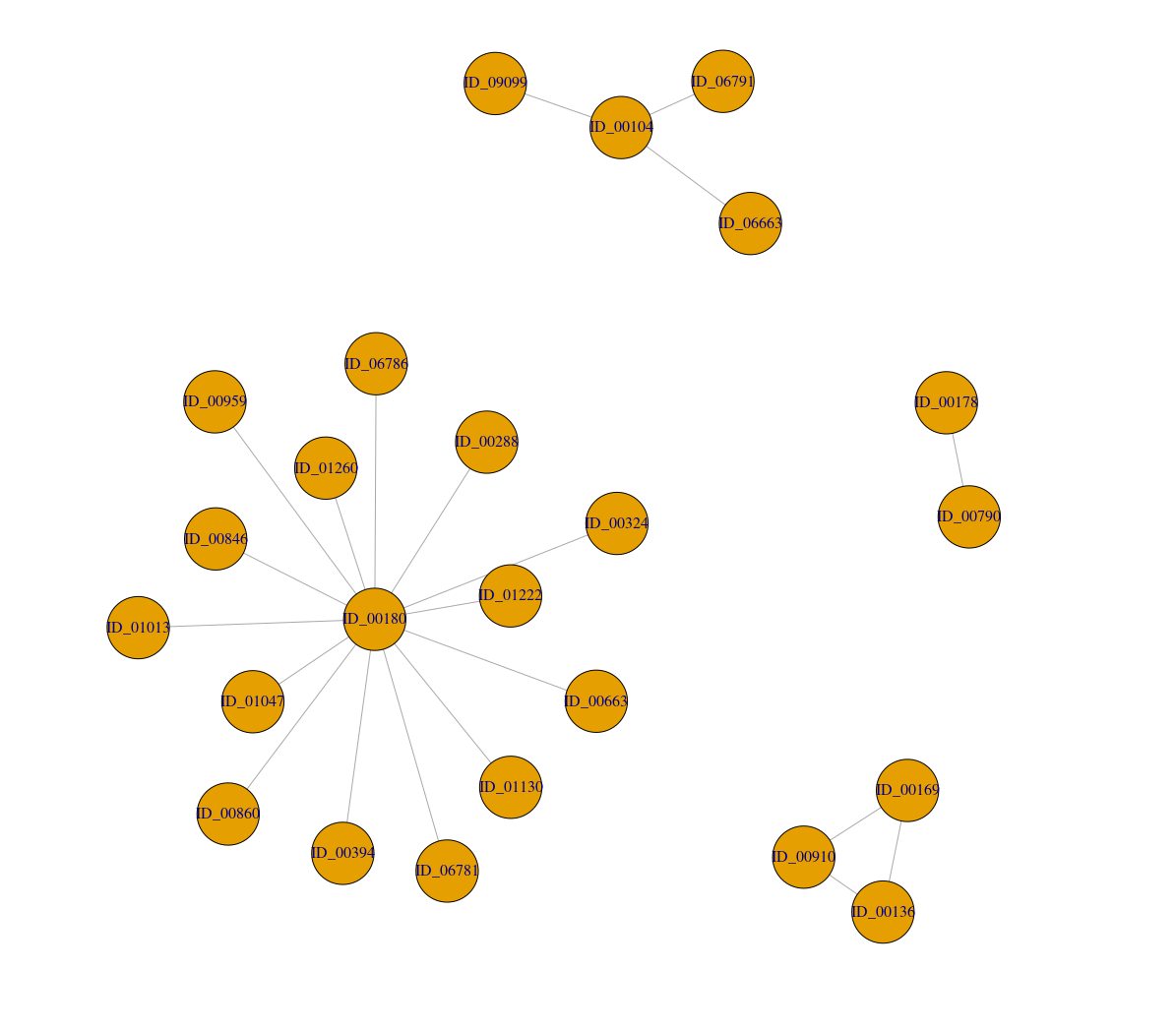
I can see the details using the below code
cl <- components(graph1)
The details are looks like
$membership
ID_00104 ID_00136 ID_00169 ID_00178 ID_00180 ID_06663 ID_06791 ID_09099 ID_00910 ID_00790 ID_01013 ID_01130 ID_01260 ID_00394 ID_00860 ID_00959 ID_01222 ID_00288 ID_00324 ID_00663 ID_00846 ID_01047 ID_06781 ID_06786
1 2 2 3 4 1 1 1 2 3 4 4 4 4 4 4 4 4 4 4 4 4 4 4
$csize
[1] 4 3 2 15
$no
[1] 4
I can also get the number of degree of a node using the below code
degree(graph1)
and the output is
ID_00104 ID_00136 ID_00169 ID_00178 ID_00180 ID_06663 ID_06791 ID_09099 ID_00910 ID_00790 ID_01013 ID_01130 ID_01260 ID_00394 ID_00860 ID_00959 ID_01222 ID_00288 ID_00324 ID_00663 ID_00846 ID_01047 ID_06781 ID_06786
3 2 2 1 14 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
I can add all components (randomly choosing 2 nodes from 2 components) using the below code (solution from one of my previous 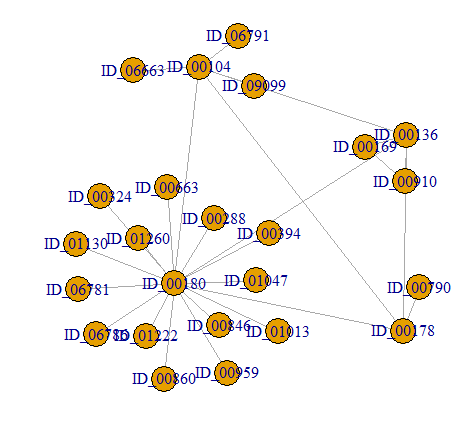
CodePudding user response:
Update
graph2 <- add.edges(
graph1,
combn(
sapply(
decompose(graph1),
function(p) sample(names(V(p))[degree(p) == max(degree(p))], 1)
), 2
),
weight = 0.01
)
plot(graph2, layout = layout_nicely(graph1))
gives
You can try
out <- combn(
decompose(graph1),
2,
FUN = function(x) {
add.edges(
graph1,
sapply(x, function(p) sample(names(V(p))[degree(p) == max(degree(p))], 1)),
weight = 0.01
)
},
simplify = FALSE
)
sapply(out,plot)