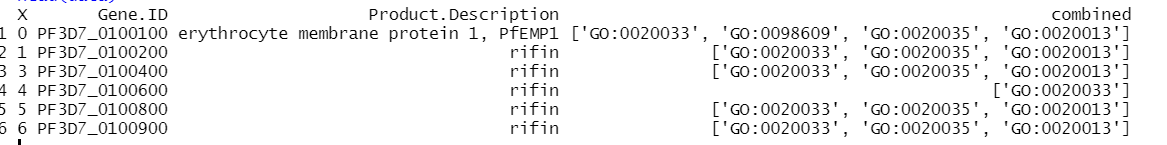
I have a csv file with one of the columns containing list data, like so,
After I read the file in R (read csv), the "combined" column has string data type. I want to be able to get it as a list. For instance, in python we would do something like,
df.combined.apply(literal_eval)
How do I do the same in R?
CodePudding user response:
Based on Konrad's answer, this is what I ended up doing:
step1= str_replace_all(text, "\\[", "")
step2= str_replace_all(step1, "\\]", "")
step3= str_replace_all(step2, "'", "")
step4= as.list(el(strsplit(step3, ",")))
PS: Thank you for pointing to the security aspect about literal_eval.
CodePudding user response:
you should be able to lapply through the column:
goid_list <- lapply(df$combined, function(x) unlist(strsplit(x, "', '")))
this gives you a list of character vectors. You will probably retain the square brackets in the first and secong GO-ID, so you might want to remove them with a gsub either within the lapply or a new lapply:
lapply(goid_list, function(x) gsub("['", "", gsub("']", "", x)))
CodePudding user response:
Just do :
library(tidyverse)
df %>%
rowwise() %>%
mutate(combined=list(reticulate::py_eval(combined)))