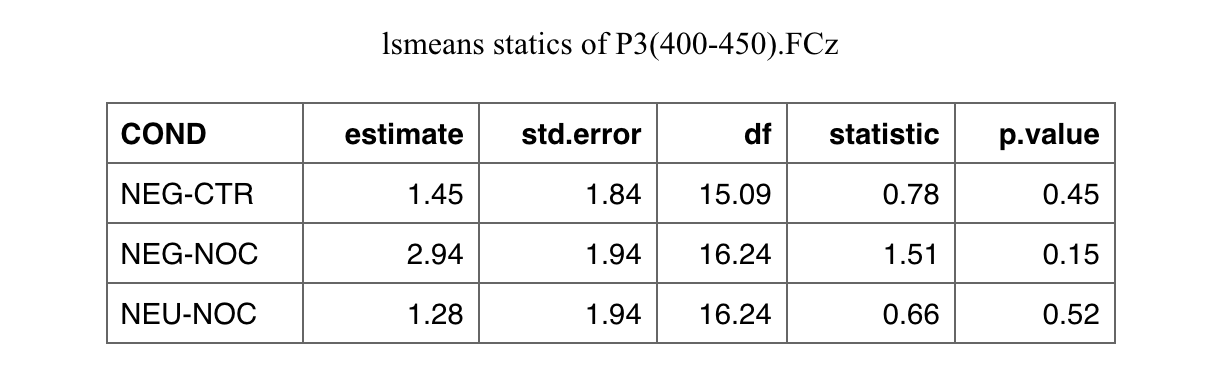
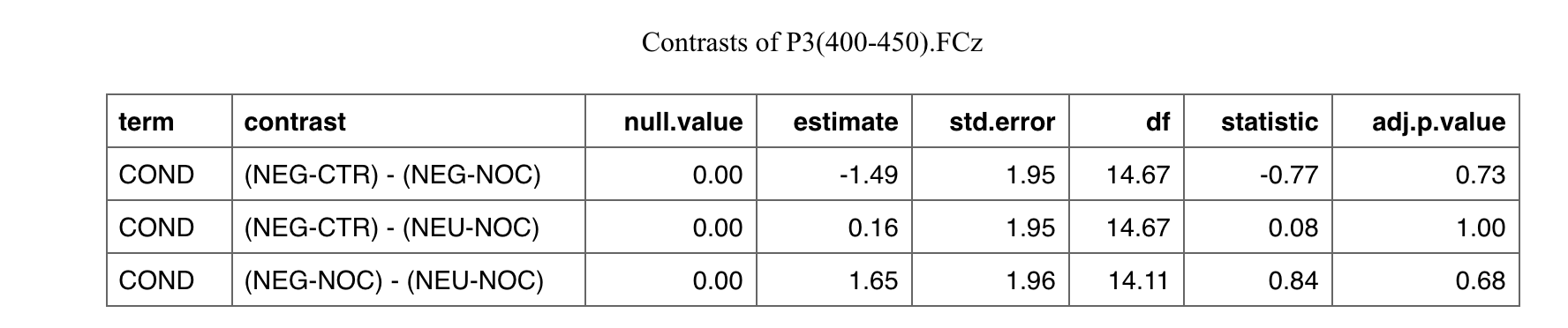
I've created a list of titles to be assigned to some tables as follows:
title <- c('P3(400-450).FCz', 'P3(400-450).Cz', 'P3(400-450).Pz',
'LPPearly(500-700).FCz', 'LPPearly(500-700).Cz',
'LPPearly(500-700).Pz', 'LPP1(500-1000).FCz',
'LPP1(500-1000).Cz', 'LPP1(500-1000).Pz',
'LPP2(1000-1500).FCz', 'LPP2(1000-1500).Cz',
'LPP2(1000-1500).Pz', 'LPP2(1000-1500).POz')
list_1 <- paste0('lsmeans statics of ', title)
list_2 <- paste0('Contrasts of ', title)
titles_tables <-append(list_1, list_2)
If I try running the following code to attribute the proper title (that actaully should return a 13 couple of tables, respectively one for lsmeans statistics and for constrasts)
tables <- md %>% map(~.x %>%
map(~broom::tidy(.x) %>% flextable::flextable() %>%
colformat_double(digits = 2) %>% theme_box() %>%
valign(valign = "center") %>% autofit() %>%
set_caption(caption = titles_tables)))
I found that each table present just the name of the first title_tables list element. Anyone knows I could sort propeprly the names to each table?
The dataset is
> dput(head(out_long, 100))
structure(list(ID = c("01", "01", "01", "04", "04", "04", "06",
"06", "06", "07", "07", "07", "08", "08", "08", "09", "09", "09",
"10", "10", "10", "11", "11", "11", "12", "12", "12", "13", "13",
"13", "15", "15", "15", "16", "16", "16", "17", "17", "17", "18",
"18", "18", "19", "19", "19", "21", "21", "21", "22", "22", "22",
"23", "23", "23", "25", "25", "25", "27", "27", "27", "28", "28",
"28", "30", "30", "30", "44", "44", "44", "46", "46", "46", "49",
"49", "49", "01", "01", "01", "04", "04", "04", "06", "06", "06",
"07", "07", "07", "08", "08", "08", "09", "09", "09", "10", "10",
"10", "11", "11", "11", "12"), GR = c("RP", "RP", "RP", "RP",
"RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP",
"RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP",
"RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP",
"RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP",
"RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP",
"RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP",
"RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP",
"RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP",
"RP", "RP", "RP", "RP", "RP", "RP", "RP", "RP"), SES = c("V",
"V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V",
"V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V",
"V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V",
"V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V",
"V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V",
"V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V",
"V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V", "V",
"V", "V", "V", "V", "V", "V", "V", "V"), COND = c("NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR", "NEG-NOC", "NEU-NOC", "NEG-CTR",
"NEG-NOC", "NEU-NOC", "NEG-CTR"), signals = c("P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz", "P3FCz",
"P3FCz", "P3FCz", "P3FCz", "P3Cz", "P3Cz", "P3Cz", "P3Cz", "P3Cz",
"P3Cz", "P3Cz", "P3Cz", "P3Cz", "P3Cz", "P3Cz", "P3Cz", "P3Cz",
"P3Cz", "P3Cz", "P3Cz", "P3Cz", "P3Cz", "P3Cz", "P3Cz", "P3Cz",
"P3Cz", "P3Cz", "P3Cz", "P3Cz"), value = c(-11.6312151716924,
-11.1438413285935, -3.99591470944713, -0.314155675382471, 0.238885648959708,
5.03749946898385, -0.213621915029167, -2.96032491743069, -1.97168681693488,
-2.83109425298642, 1.09291198163802, -6.692991645215, 4.23849942428043,
2.9898889629932, 3.5510699900835, 9.57481668808606, 5.4167795618285,
1.7067607715475, -6.13036076093477, -2.82955734597919, -2.50672211111696,
0.528517585832501, 8.16418133488309, 1.88777321897925, -7.73588468896919,
-9.83058052401056, -6.97442700196932, 1.27327945355082, 2.11962397764132,
0.524299677616254, -1.83310726842883, 0.658810483381172, -0.261373488428192,
4.37524298634374, 0.625555654900511, 3.19617639836154, 0.0405517582137798,
-3.29357103412113, -0.381435057304614, -5.73445509910268, -6.1129152355645,
-2.45744234877604, 2.95352732001065, 0.527721249096473, 1.91803490989119,
-3.46703346467546, -2.40438419043702, -5.35374408162217, -7.27028665849262,
-7.1532211375959, -5.39955520296854, 2.65765002364624, 0.372495441513391,
6.24433066412776, 1.85698518142405, -0.564454675803529, -0.068523080368053,
-7.04782633579147, -4.52263283590558, -6.62134671432544, 4.56661945182626,
3.05859761335498, 2.02997952225347, -6.10523962206958, -0.521871236969702,
-3.97851995684846, -2.61258020387919, -4.13974828699279, -3.9210032516844,
-4.63162466544638, -4.36762718685405, -6.71005969834916, -4.22719611676328,
-0.229916506217565, -5.69725200870146, -5.16524399006139, -5.53112490175437,
0.621502123415388, 2.23100741241039, 3.96990710862955, 7.75899775608441,
-1.30019374375434, -3.59899040898949, -1.92340529575071, 2.19344184533265,
5.87900720863083, -5.92378937757888, 2.44958531767688, 3.10043497883256,
1.65779442628225, 13.7118233181713, 6.86178446511352, 5.31481098188172,
-4.13240668697805, 0.162182285588285, 0.142083484505352, 5.42592103255673,
14.5496375672716, 4.52018125654081, -2.40677805475299)), row.names = c(NA,
-100L), class = c("tbl_df", "tbl", "data.frame"))
>
Here the way I got the lsmeans statistics, which the tables are referring to:
md1 <- out_long %>%
group_by(signals) %>%
do(fit = lmerTest::lmer(value ~ COND (1 |ID), data = .)) %>%
pull(fit) %>%
lapply(., function(m) lsmeans(m, pairwise ~ COND, adjust="tukey"))
CodePudding user response:
Consider changing the append to
titles_tables <- Map(c, list_1, list_2)
as append does concatenation of the two lists into a vector, whereas we need to name the nested list elements with each corresponding elements of list_1 and list_2. So, it is better to keep it in a list of vectors by concatenating the corresponding elements.
and then use map2 (as mentioned in the comments). Here, it is a nested list. So, we need two map2. i.e. the first map2, gets the 'md1' and the corresponding list of titles_tables, and within the next map2, it will be looping over the corresponding individual elements
library(dplyr)
library(purrr)
library(flextable)
library(emmeans)
output <- md1 %>%
map2(titles_tables[seq_along(md1)], ~{
title <- .y
.x %>%
map2(title, ~broom::tidy(.x) %>% flextable::flextable() %>%
colformat_double(digits = 2) %>% theme_box() %>%
valign(valign = "center") %>% autofit() %>%
set_caption(caption = .y))
})
-check the output
output[[1]]$lsmeans
output[[1]]$contrasts