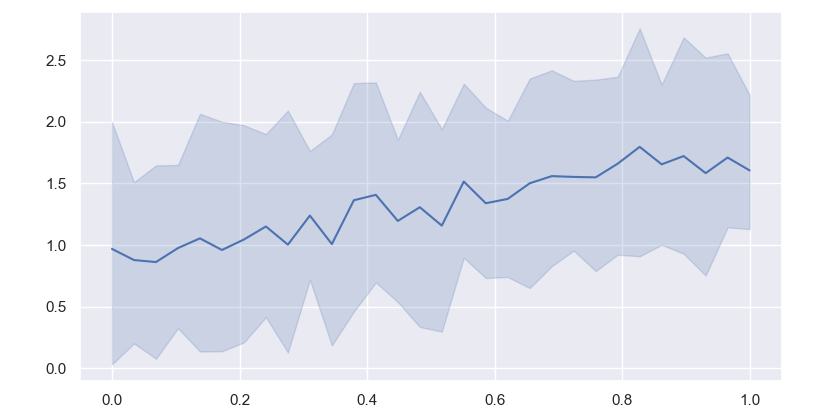
I have some data stored as [samples, x_values_toconsider] and then I give it to seaborn. I've tried many combinations for it to plot the error bands but it doesn't seem to do it, it simply plots each of my samples for a specific feature/value of x. So it ends up have samples number of curves, which is NOT what I want. I've also tried to arrange it as a data frame but that didn't help either.
Why is seaborn plotting each individual sample as it's own curve? I want it to aggregate it with the usual confidence intervals.
Self contained Reproducible code:
#%%
"""
https://seaborn.pydata.org/tutorial/relational.html#relational-tutorial
https://seaborn.pydata.org/examples/errorband_lineplots.html
https://www.youtube.com/watch?v=G3F0EZcW9Ew
https://github.com/knathanieltucker/seaborn-weird-parts/commit/3e571fd8e211ea04b6c9577fd548e7e532507acf
https://github.com/knathanieltucker/seaborn-weird-parts/blob/3e571fd8e211ea04b6c9577fd548e7e532507acf/tsplot.ipynb
"""
from collections import OrderedDict
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
from pandas import DataFrame
import pandas as pd
print(sns)
np.random.seed(22)
sns.set(color_codes=True)
# the number of x values to consider in a given range e.g. [0,1] will sample 10 raw features x sampled at in [0,1] interval
num_x: int = 10
# the repetitions for each x feature value e.g. multiple measurements for sample x=0.0 up to x=1.0 at the end
rep_per_x: int = 5
total_size_data_set: int = num_x * rep_per_x
print(f'{total_size_data_set=}')
# - create fake data set
# only consider 10 features from 0 to 1
x = np.linspace(start=0.0, stop=1.0, num=num_x)
# to introduce fake variation add uniform noise to each feature and pretend each one is a new observation for that feature
noise_uniform: np.ndarray = np.random.rand(rep_per_x, num_x)
# same as above but have the noise be the same for each x (thats what the 1 means)
noise_normal: np.ndarray = np.random.randn(rep_per_x, 1)
# signal function
sin_signal: np.ndarray = np.sin(x)
# [rep_per_x, num_x]
data: np.ndarray = sin_signal noise_uniform noise_normal
# data_od: OrderedDict = OrderedDict()
# for idx_x in range(num_x):
# # [rep_per_x, 1]
# samples_for_x: np.ndarray = data[:, idx_x]
# data_od[str(x[idx_x])] = samples_for_x
#
# data_df = pd.DataFrame(data_od)
# data = data_df
print(data)
ax = sns.lineplot(data=data)
# ax = sns.lineplot(data=data, err_style='band')
# ax = sns.lineplot(data=data, err_style='bars')
# ax = sns.lineplot(data=data, ci='sd', err_style='band')
# ax = sns.lineplot(data=data, ci='sd', err_style='bars')
# ax = sns.relplot(data=data)
plt.show()
#%%
"""
https://seaborn.pydata.org/examples/errorband_lineplots.html
"""
# import numpy as np
# import seaborn as sns
# from matplotlib import pyplot as plt
# from pandas import DataFrame
#
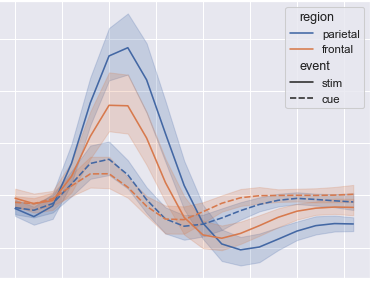
# fmri: DataFrame = sns.load_dataset("fmri")
# print(fmri)
# sns.lineplot(x="timepoint", y="signal", hue="region", style="event", data=fmri)
# plt.show()
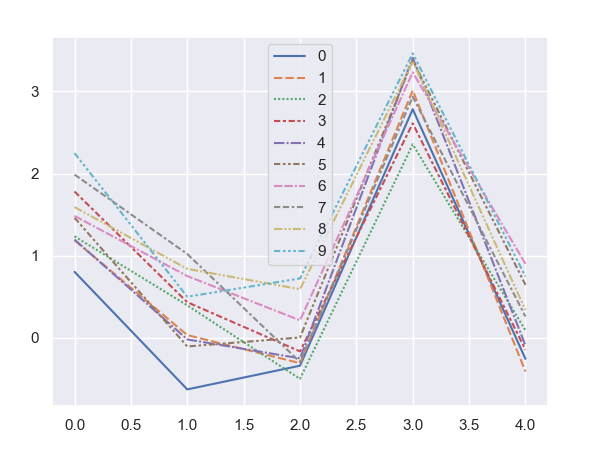
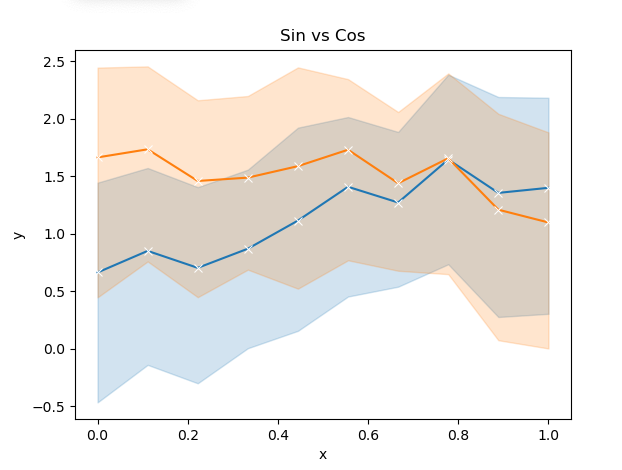
wrong plot produce
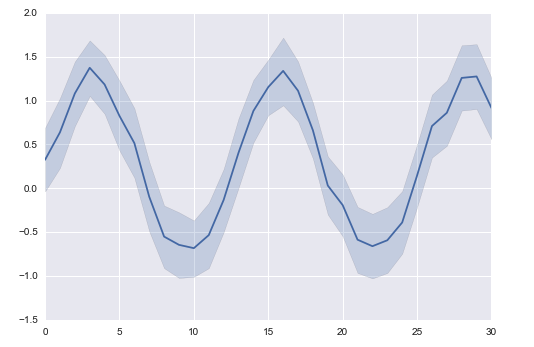
but hoped for something like this (and with addition lines with their own error bands would be even better cuz I have many matrices!)
like:
Note that I do not want to pre-calculate the stds and plot the bands so most of those questions/answers don't work for me.
What puzzles me is that when I pass it the fmri data it works but not when I pass it my matrix of observations for each x value...
related posts:
Note that the tutorial examples where no explicit
xis given, work with data in the form of a pandas dataframe, or a column of such in dataframe. In that case, the index of the dataframe can be used for the x-values.Or you could concatenate all the values. Working with a dataframe might make it easier to see what's going on, and would also automatically use the column names to label the axes and the legend.
ax = sns.lineplot(x=np.tile(x, rep_per_x1 rep_per_x2), y=np.concatenate([data1.ravel(), data2.ravel()]), hue=np.concatenate([np.repeat(1, num_x * rep_per_x1), np.repeat(2, num_x * rep_per_x2)]), palette=['crimson', 'dodgerblue'])CodePudding user response:
For the sake of having a full example with reusable code for future users:
def plot_seaborn_curve_with_x_values_y_values(x: np.ndarray, y: np.ndarray, xlabel: str, ylabel: str, title: str, curve_label: Optional[str] = None, err_style: str = 'band', marker: Optional[str] = 'x', dashes: bool = False, show: bool = False ): """ Given a list of x values in a range with num_x_values number of x values in that range and the corresponding samples for each specific x value (so [samples_per_x] for each value of x giving in total a matrix of size [samples_per_x, num_x_values]), plot aggregates of them with error bands. Note that the main assumption is that each x value has a number of y values corresponding to it (likely due to noise for example). Note: - if you want string in the specific x axis point do sns.lineplot(x=np.tile([f'Layer{i}' for i in range(1, num_x 1)], rep_per_x),...) assuming the x values are the layers. https://stackoverflow.com/questions/69888181/how-to-show-error-bands-for-pure-matrices-samples-x-range-with-seaborn-error/69889619?noredirect=1#comment123544763_69889619 - note you can all this function multiple times to insert different curves to your plot. - note its recommended call show only for if you have one or at the final curve you want to add. - if you want bands and bars it might work if you call this function twice but using the bar and band argument for each call. ref: - https://stackoverflow.com/questions/69888181/how-to-show-error-bands-for-pure-matrices-samples-x-range-with-seaborn-error/69889619?noredirect=1#comment123544763_69889619 :param x: [num_x_values] :param y: [samples_per_x, num_x_values] :param xlabel: :param ylabel: :param title: :param curve_label: :param err_style: :param marker: :param dashes: :param show: :return: """ import seaborn as sns samples_per_x: int = y.shape[0] num_x_values: int = x.shape[0] assert(num_x_values == y.shape[1]), f'We are plotting aggreagates for one specific value of x multple values of y,' \ f'thus we need to have the same number of x values match in the x axis.' # - since seaborn expects a an x value paired with it's y value, let's flatten the y's and make sure the corresponding # x value is aligned with it's y value [num_x_values * samples_per_x] x: np.ndarray = np.tile(x, samples_per_x) # np.tile = Construct an array by repeating A the number of times given by reps. assert (x.shape == (num_x_values * samples_per_x,)) y: np.ndarray = np.ravel(y) # flatten the y's to match the x values to have the x to it's corresponding y assert (y.shape == (num_x_values * samples_per_x,)) assert (x.shape == y.shape) # - plot ax = sns.lineplot(x=x, y=y, err_style=err_style, label=curve_label, marker=marker, dashes=dashes) plt.title(title) plt.xlabel(xlabel) plt.ylabel(ylabel) if show: plt.show()e.g.
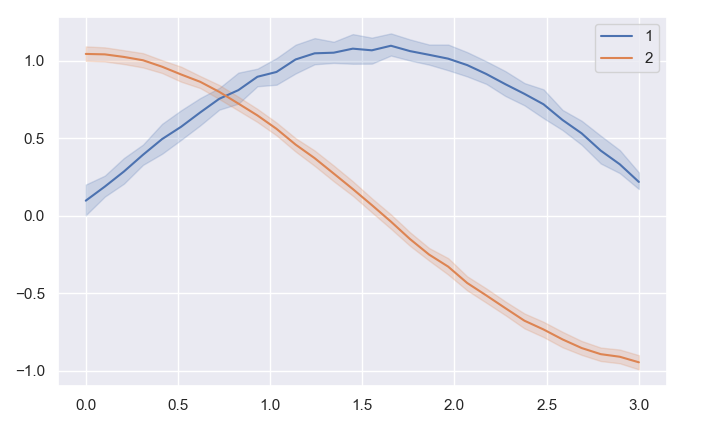
def plot_seaborn_curve_with_x_values_y_values_test(): # the number of x values to consider in a given range e.g. [0,1] will sample 10 raw features x sampled at in [0,1] interval num_x: int = 10 # the repetitions for each x feature value e.g. multiple measurements for sample x=0.0 up to x=1.0 at the end rep_per_x: int = 5 total_size_data_set: int = num_x * rep_per_x print(f'{total_size_data_set=}') # - create fake data set # only consider 10 features from 0 to 1 x = np.linspace(start=0.0, stop=1.0, num=num_x) # to introduce fake variation add uniform noise to each feature and pretend each one is a new observation for that feature noise_uniform: np.ndarray = np.random.rand(rep_per_x, num_x) # same as above but have the noise be the same for each x (thats what the 1 means) noise_normal: np.ndarray = np.random.randn(rep_per_x, 1) # signal function sin_signal: np.ndarray = np.sin(x) cos_signal: np.ndarray = np.cos(x) # [rep_per_x, num_x] y1: np.ndarray = sin_signal noise_uniform noise_normal y2: np.ndarray = cos_signal noise_uniform noise_normal plot_seaborn_curve_with_x_values_y_values(x=x, y=y1, xlabel='x', ylabel='y', title='Sin vs Cos') plot_seaborn_curve_with_x_values_y_values(x=x, y=y2, xlabel='x', ylabel='y', title='Sin vs Cos') plt.show()output: