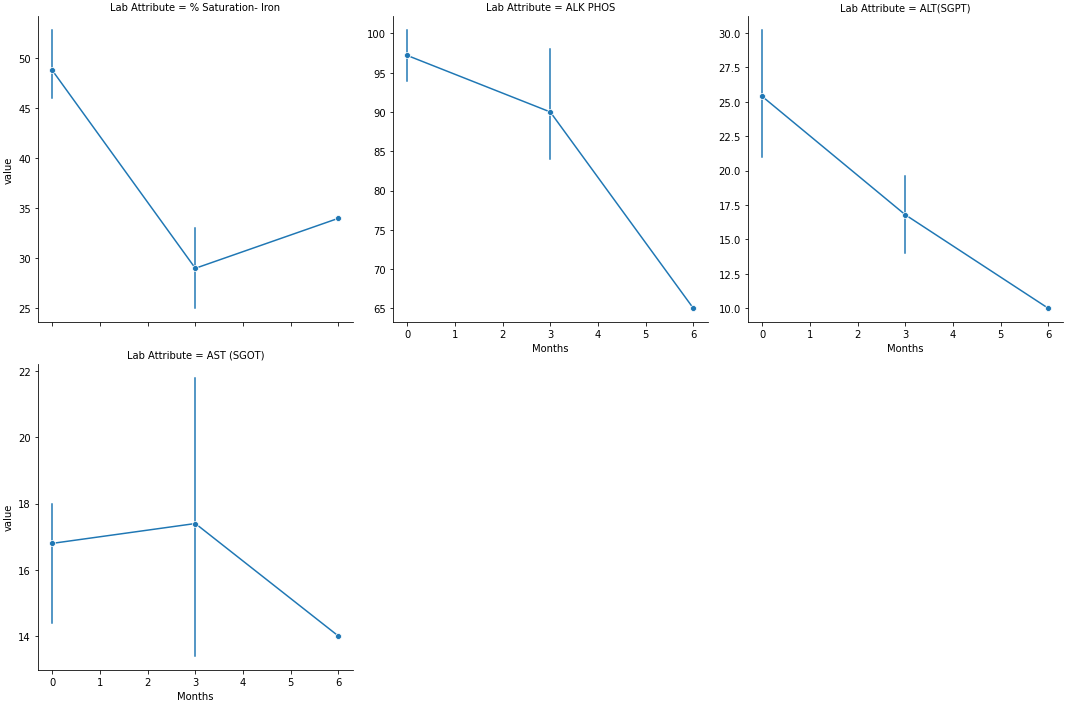
Using this code I created a seaborn plot to visualize multiple variables in a long format dataset.
import pandas as pd
import seaborn as sns
data = {'Patient ID': [11111, 11111, 11111, 11111, 22222, 22222, 22222, 22222, 33333, 33333, 33333, 33333, 44444, 44444, 44444, 44444, 55555, 55555, 55555, 55555],
'Lab Attribute': ['% Saturation- Iron', 'ALK PHOS', 'ALT(SGPT)', 'AST (SGOT)', '% Saturation- Iron', 'ALK PHOS', 'ALT(SGPT)', 'AST (SGOT)', '% Saturation- Iron', 'ALK PHOS', 'ALT(SGPT)', 'AST (SGOT)', '% Saturation- Iron', 'ALK PHOS', 'ALT(SGPT)', 'AST (SGOT)', '% Saturation- Iron', 'ALK PHOS', 'ALT(SGPT)', 'AST (SGOT)'],
'Baseline': [46.0, 94.0, 21.0, 18.0, 56.0, 104.0, 31.0, 12.0, 50.0, 100.0, 33.0, 18.0, 46.0, 94.0, 21.0, 18.0, 46.0, 94.0, 21.0, 18.0],
'3 Month': [33.0, 92.0, 19.0, 25.0, 33.0, 92.0, 21.0, 11.0, 33.0, 102.0, 18.0, 17.0, 23.0, 82.0, 13.0, 17.0, 23.0, 82.0, 13.0, 17.0],
'6 Month': [34.0, 65.0, 10.0, 14.0, 34.0, 65.0, 10.0, 14.0, 34.0, 65.0, 10.0, 14.0, 34.0, 65.0, 10.0, 14.0, 34.0, 65.0, 10.0, 14.0]}
df = pd.DataFrame(data)
# reshape the dataframe
dfm = df_labs.melt(id_vars=['Patient_ID', 'Lab_Attribute'], var_name='Months')
# change the Months values to numeric
dfm.Months = dfm.Months.map({'Baseline': 0, '3 Month': 3, '6 Month': 6})
# plot a figure level line plot with seaborn
p = sns.relplot(data=dfm, col='Lab_Attribute', x='Months', y='value', hue='Patient_ID', kind='line', col_wrap=5, marker='o', palette='husl',facet_kws={'sharey': False, 'sharex': True},err_style="bars", ci=95,)
plt.savefig('gmb_nw_labs.jpg')
The plots work great, though for some reason the error bars are not displaying, even after adding:
err_style="bars", ci=95,
to sns.replot()
p = sns.relplot(data=dfm, col='Lab_Attribute', x='Months', y='value', hue='Patient_ID', kind='line', col_wrap=5, marker='o', palette='husl',facet_kws={'sharey': False, 'sharex': True},err_style="bars", ci=95,)
Can anyone tell me why this is, are there maybe just too few data points in my data set?
CodePudding user response:
- Each datapoint is separated by
hue, so there are no error bars because no data is being combined. Removehue='Patient ID'to only show the mean line and error bars. - Alternatively,
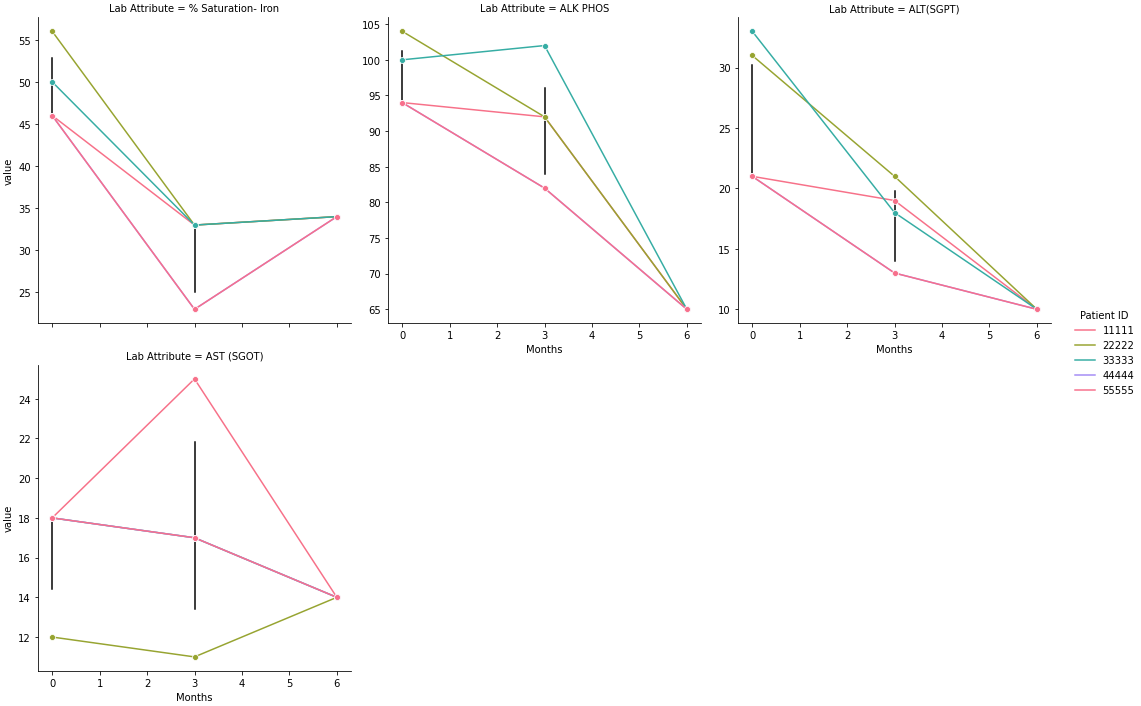
- Original implementation without
hue='Patient ID'
p = sns.relplot(data=dfm, col='Lab Attribute', x='Months', y='value', kind='line', col_wrap=3, marker='o', palette='husl', facet_kws={'sharey': False, 'sharex': True}, err_style="bars", ci=95) - Original implementation without