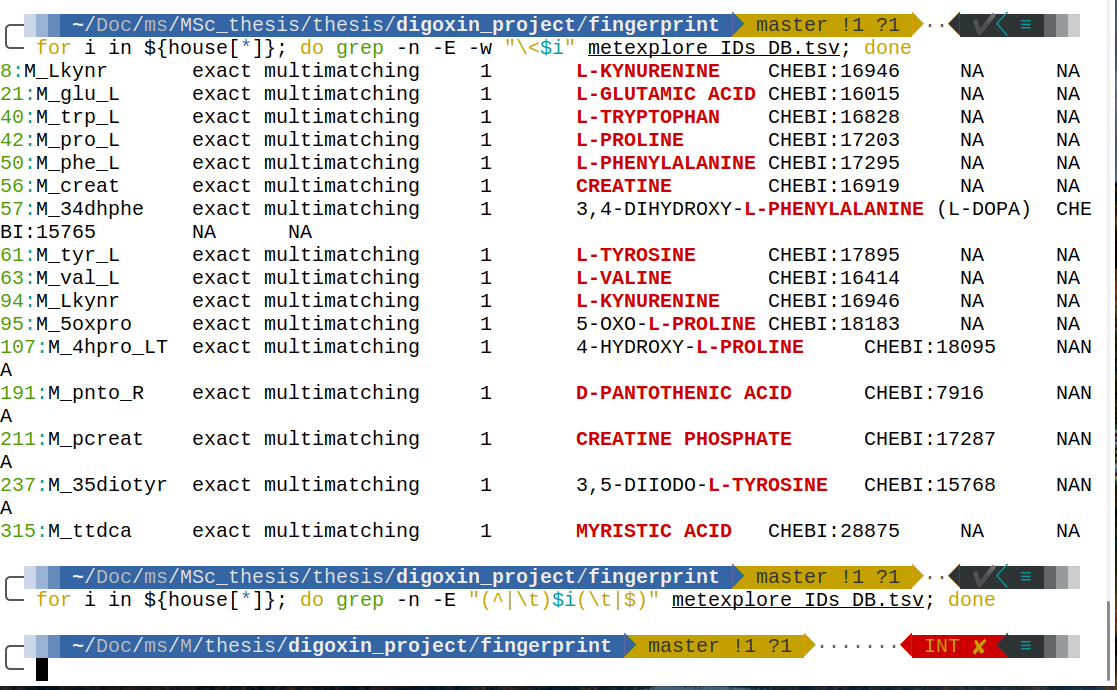
I have an list/array in zsh which is house=$(cat corrected_inhouse_list.txt)
Containing:
N-METHYL-L-GLUTAMIC ACID
L-GLUTAMIC ACID
CREATINE
L-PROLINE
CREATINE PHOSPHATE
L-VALINE
L-TYROSINE
L-KYNURENINE
L-PHENYLALANINE
PHENYLETHANOLAMINE
D-PANTOTHENIC ACID
L-TRYPTOPHAN
MYRISTIC ACID
File "metexplore_ID.tsv":
8:M_Lkynr exact multimatching 1 L-KYNURENINE CHEBI:16946 NA NA
21:M_glu_L exact multimatching 1 L-GLUTAMIC ACID CHEBI:16015 NA NA
40:M_trp_L exact multimatching 1 L-TRYPTOPHAN CHEBI:16828 NA NA
42:M_pro_L exact multimatching 1 L-PROLINE CHEBI:17203 NA NA
50:M_phe_L exact multimatching 1 L-PHENYLALANINE CHEBI:17295 NA NA
56:M_creat exact multimatching 1 CREATINE CHEBI:16919 NA NA
57:M_34dhphe exact multimatching 1 3,4-DIHYDROXY-L-PHENYLALANINE (L-DOPA) CHEBI:15765 NA NA
61:M_tyr_L exact multimatching 1 L-TYROSINE CHEBI:17895 NA NA
63:M_val_L exact multimatching 1 L-VALINE CHEBI:16414 NA NA
94:M_Lkynr exact multimatching 1 L-KYNURENINE CHEBI:16946 NA NA
95:M_5oxpro exact multimatching 1 5-OXO-L-PROLINE CHEBI:18183 NA NA
107:M_4hpro_LT exact multimatching 1 4-HYDROXY-L-PROLINE CHEBI:18095 NANA
171:M_pcreat exact multimatching 1 PHOSPHOCREATINE CHEBI:17287 NA NA
191:M_pnto_R exact multimatching 1 D-PANTOTHENIC ACID CHEBI:7916 NANA
211:M_pcreat exact multimatching 1 CREATINE PHOSPHATE CHEBI:17287 NANA
237:M_35diotyr exact multimatching 1 3,5-DIIODO-L-TYROSINE CHEBI:15768 NANA
315:M_ttdca exact multimatching 1 MYRISTIC ACID CHEBI:28875
And I want to use grep to match this words on a file. The Problem is what one can see in the picture, grep also captures words that contain but not start with my word of interes.
I've tried:
for i in ${house[*]}; do grep -n -E "^\s*\{$i}\>" metexplore_IDs_DB.tsv; done
for i in ${house[*]}; do grep -n -E -w "\<$i" metexplore_IDs_DB.tsv; done
for i in ${house[*]}; do grep -n -E "(^|\t)$i" metexplore_IDs_DB.tsv; done
What can I do to achieve my goal? the desired output would be without lines 57, 95, 107 and 237.
CodePudding user response:
You may comside this awk that builds regex for each entry from your list and then searches that regex anywhere in the csv file:
awk 'NR==FNR {
kw[ "(^|[[:blank:]])" $0 "([[:blank:]]|$)" ]
next
}
{
for (w in kw)
if ( $0 ~ w ) {
print
next
}
}' words.lst file.csv
i8:M_Lkynr exact multimatching 1 L-KYNURENINE CHEBI:16946 NA NA
21:M_glu_L exact multimatching 1 L-GLUTAMIC ACID CHEBI:16015 NA NA
40:M_trp_L exact multimatching 1 L-TRYPTOPHAN CHEBI:16828 NA NA
42:M_pro_L exact multimatching 1 L-PROLINE CHEBI:17203 NA NA
50:M_phe_L exact multimatching 1 L-PHENYLALANINE CHEBI:17295 NA NA
56:M_creat exact multimatching 1 CREATINE CHEBI:16919 NA NA
61:M_tyr_L exact multimatching 1 L-TYROSINE CHEBI:17895 NA NA
63:M_val_L exact multimatching 1 L-VALINE CHEBI:16414 NA NA
94:M_Lkynr exact multimatching 1 L-KYNURENINE CHEBI:16946 NA NA
191:M_pnto_R exact multimatching 1 D-PANTOTHENIC ACID CHEBI:7916 NANA
211:M_pcreat exact multimatching 1 CREATINE PHOSPHATE CHEBI:17287 NANA
315:M_ttdca exact multimatching 1 MYRISTIC ACID CHEBI:28875
CodePudding user response:
It looks like you're always matching field 3. So awk would be a better solution, since you can simply do an exact match with the whole field:
for i in "${house[@]}"; do
awk -F'\t' -v i="$i" '$3 == i' metexplore_IDs_DB.tsv
done
Don't forget the quotes around ${house[@]}; otherwise elements like L-GLUTAMIC ACID will be treated as two different words to match.
You can also avoid creating the array and looping by loading corrected_inhouse_list.txt directly into an awk array:
awk -F'\t' -v i="$i" '
NR == FNR '{houses[$0] ; next}
$5 in houses' corrected_inhouse_list.txt metexplore_IDs_DB.tsv