I would like to read out specific columns from multiple (100s) CSV files to create a new CSV file. The column header will be renamed with the source filename where the column is extracted. I have written some lines of code to do this, however, I have been unable to effectively rename the column header without the extension (See attachment). I will appreciate some help. Thanks a lot.
import os
import csv
import pandas as pd
folder = "C:/Users/Doc/Data"
files = os.scandir(folder)
E2080 = []
with os.scandir(folder) as files:
for file in files:
#print(file)
df = pd.read_csv(file, index_col=None)
dist = {file: (df['lnt_dist'])}
E = pd.DataFrame(dist)
E2080.append(E)
dist = pd.concat(E2080, ignore_index=False, axis=1)
dist.head()
dist.to_csv('E2080', index=False)
CodePudding user response:
You should use file.name instead of file to get string with name.
And with string you can use .split(".") to get name without extension.
for file in os.scandir(folder):
print(file.name, '=>', file.name.split(".")[0])
Or you could use pathlib.Path instead of os.scandir() to have more functions.
for file in pathlib.Path('test').iterdir():
print(file.name, '=>', file.stem)
CodePudding user response:
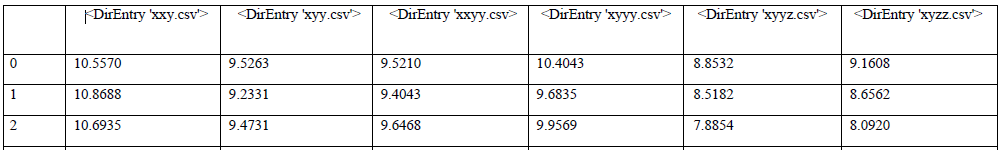
This is the final code that worked for me (see output 1):
E2080 = []
with os.scandir(folder) as files:
for file in files:
#print(file)
df = pd.read_csv(file, index_col=None)
dist = {file: (df['lnt_dist'])}
E = pd.DataFrame(dist)
E_1 = E.rename(columns={file: file.name.split('.')[0]}) # rename df header while dropping the ext **[.csv]** and the `os.scandir` attribute `<DirEntry>`
E2080.append(E_1)
dist = pd.concat(E_28, ignore_index=False, axis=1)
#dist.head()
dist.to_csv('E2080.csv', index=False)
