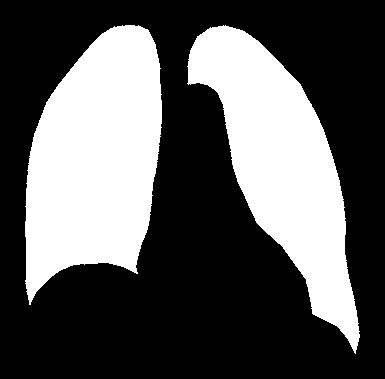
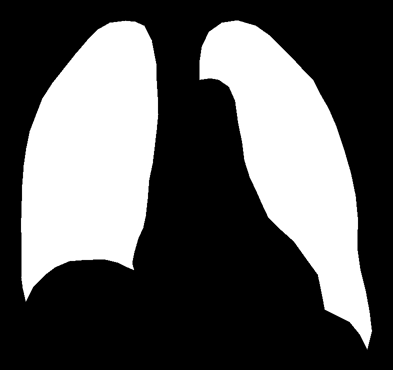
I have merged two lung images using an online merging tool to display a full image.
The problem is that since the background of each image is black I got an unwanted gap between the lungs in the image displayed.
I would like to know if there is a way to remove an area from an image either with code with an algorithm or with an online tool and reduce the gap between the lungs.
Another approach I checked was using OpenCV with Python for a panoramic image stitching, which I will try as a last resort to connect my images.
My desired result:
CodePudding user response:
Here is one way as mention in my comments for Python/OpenCV.
- Read input and convert to grayscale
- Threshold to binary
- Get external contours
- Filter contours to keep only large ones and put bounding box values into list. Also compute the max width and max height from bounding boxes.
- Set desired amount of padding
- Create a black image the size of max width and max height
- Sort the bounding box list by x value
- Get the first item from the list and crop and pad it
- Create a black image the size of max height and desired pad
- Horizontally concatenate with previous padded crop
- Get the second item from the list and crop and pad it
- Horizontally concatenate with previous padded result
- Pad all around as desired
- Save the output
Input:
import cv2
import numpy as np
# load image
img = cv2.imread("lungs_mask.png", cv2.IMREAD_GRAYSCALE)
# threshold
thresh = cv2.threshold(img, 128, 255, cv2.THRESH_BINARY)[1]
# get the largest contour
contours = cv2.findContours(thresh, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
contours = contours[0] if len(contours) == 2 else contours[1]
# get bounding boxes of each contour if area large and put into list
cntr_list=[]
max_ht = 0
max_wd = 0
for cntr in contours:
area = cv2.contourArea(cntr)
if area > 10000:
x,y,w,h = cv2.boundingRect(cntr)
cntr_list.append([x,y,w,h])
if h > max_ht:
max_ht = h
if w > max_wd:
max_wd = w
# set padded
padding = 25
# create black image of size max_wd,max_ht
black = np.zeros((max_ht,max_wd), dtype=np.uint8)
# sort contours by x value
def takeFirst(elem):
return elem[0]
cntr_list.sort(key=takeFirst)
# Take first entry in sorted list and crop and pad
item = cntr_list[0]
x = item[0]
y = item[1]
w = item[2]
h = item[3]
crop = thresh[y:y h, x:x w]
result = black[0:max_ht, 0:w]
result[0:h, 0:w] = crop
# create center padding and concatenate
pad_center_img = np.zeros((max_ht,padding), dtype=np.uint8)
result = cv2.hconcat((result, pad_center_img))
# Take second entry in sorted list and crop, pad and concatenate
item = cntr_list[1]
x = item[0]
y = item[1]
w = item[2]
h = item[3]
crop = thresh[y:y h, x:x w]
temp = black[0:max_ht, 0:w]
temp[0:h, 0:w] = crop
result = cv2.hconcat((result, temp))
# Pad all around as desired
result = cv2.copyMakeBorder(result, 25, 25, 25, 25, borderType=cv2.BORDER_CONSTANT, value=(0))
# write result to disk
cv2.imwrite("lungs_mask_cropped.jpg", result)
# display it
cv2.imshow("thresh", thresh)
cv2.imshow("result", result)
cv2.waitKey(0)
Result:
CodePudding user response:
The Concept:
-
Notes:
- At the line:
lung_1 = cv2.copyMakeBorder(img[y1: y1 h2, x1: x1 w1], 20, 20, 20, 20, cv2.BORDER_CONSTANT)notice that we used
h2instead ofh1, as if we were to use theh1we defined, thenp.hstack()method would throw an error due to the different heights of the arrays.- The
sorted()at
(x1, y1, w1, h1), (x2, y2, w2, h2) = sorted(map(cv2.boundingRect, contours))is to sort the
x, y, w, hby theirxproperty, so that the lungs are concatenated from left to right.