If I want to plot an empirical density, I would go:
library(ggplot2) ggplot() geom_density(aes(x = rbeta(100,3,1)))
or
library(ggplot2)
ggplot()
geom_histogram(aes(x = rbeta(100,3,1)))
where rbeta(100,3,1) can be any vector.
If I want to plot a theoretical density, I could go:
library(ggplot2)
ggplot(data = data.frame(x = c(0, 1)), mapping = aes(x = x))
stat_function(fun = dbeta, args = c(3,1), n = 100)
But when I try to plot the first curve over the second:
library(ggplot2)
ggplot(data = data.frame(x = c(0, 1)), mapping = aes(x = x))
stat_function(fun = dbeta, args = c(3,1), n = 100)
geom_histogram(aes(x = rbeta(100,3,1)))
I will get an error.
How can I plot an empirically determined density over a theoretical?
CodePudding user response:
This seems to work.
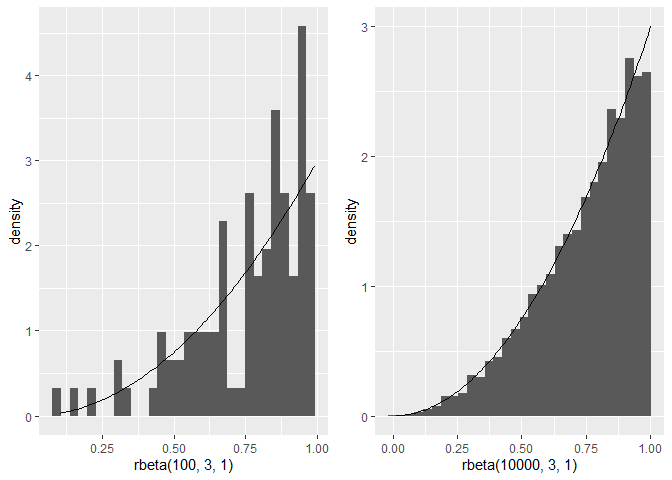
Plot the histogram first, then the density. And with more data points the histogram fits the theoretical density better.
library(ggplot2)
library(gridExtra)
set.seed(2022)
p1 <- ggplot()
geom_histogram(aes(x = rbeta(100,3,1), y = ..density..), bins = 30)
stat_function(fun = dbeta, args = c(3,1), n = 100)
p2 <- ggplot()
geom_histogram(aes(x = rbeta(10000,3,1), y = ..density..), bins = 30)
stat_function(fun = dbeta, args = c(3,1), n = 100)
grid.arrange(p1, p2, ncol=2)
Created on 2022-02-20 by the reprex package (v2.0.1)
