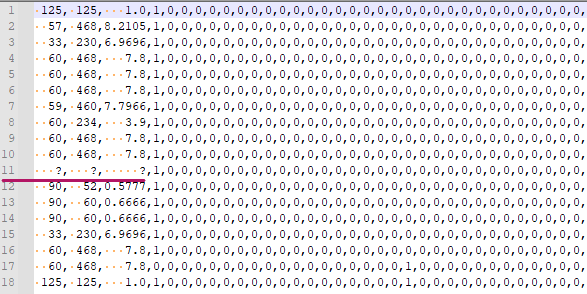
I have a dataset with a customized missing values which is the character `\?` but a cell with the missing value also contains whitespaces with inconsistent number of space characters. As in my example picture, at row 11, It could have 3 spaces, or 4 spaces.
So my idea is to apply the str.strip() function for each cell to identify it as the missing values and drop it, but it still is not recognized as missing values.
df = pd.read_csv('full_name', header=None, na_values=['?'])
df = df.apply(lambda x: x.str.strip() if x.dtype== 'object' else x)
df.dropna(axis=0, inplace=True, how='any')
df.head(20)]
what is an efficient way to solve this?
CodePudding user response:
Use:
df = pd.DataFrame({'test': [1,2, ' ? ', ' ? ']})
df[~df['test'].str.contains('\?', na=False)]
CodePudding user response:
dropna drops NaN values. Since your NaNs are actually ?, you could replace them with NaN and use dropna:
df = df.replace('?', np.nan).dropna()
mask them and use dropna:
df = df.mask(df.eq('?')).dropna()
or simply filter those rows out and only select rows without any ?:
df = df[df.ne('?').all(axis=1)]