I want to achieve the same end goal as this question: Create a single heatmap based on two symmetric matrices in R but to take it further than the answer currently provided.
The answer given does not explain how one would go about having different colours for the upper and lower sections of the matrix and different scales?
Here is the example dataset:
library(Matrix)
set.seed(123)
s1<-forceSymmetric(matrix(round(rnorm(25),2),5))
colnames(s1)<-LETTERS[1:5]
rownames(s1)<-LETTERS[6:10]
diag(s1)<-1
s2<-forceSymmetric(matrix(round(rbinom(25,25,0.3),2),5))
colnames(s2)<-LETTERS[1:5]
rownames(s2)<-LETTERS[6:10]
diag(s2)<-1
s1
# 5 x 5 Matrix of class "dsyMatrix"
# A B C D E
# F 1.00 1.72 1.22 1.79 -1.07
# G 1.72 1.00 0.36 0.50 -0.22
# H 1.22 0.36 1.00 -1.97 -1.03
# I 1.79 0.50 -1.97 1.00 -0.73
# J -1.07 -0.22 -1.03 -0.73 1.00
s2
# 5 x 5 Matrix of class "dsyMatrix"
# A B C D E
# F 1 6 8 7 9
# G 6 1 5 9 8
# H 8 5 1 10 9
# I 7 9 10 1 1
# J 9 8 9 1 1
The suggested answer suggests to add the two matrices together as so:
#Get upper diagonal
reverse = s1[,ncol(s1):1]
diag(reverse) = 0
reverse[lower.tri(reverse, diag = FALSE)] <- 0
upper = reverse[,ncol(reverse):1]
# Get lower diagonal
reverse1 = s2[,ncol(s2):1]
diag(reverse1) = 0
reverse1[upper.tri(reverse1, diag = FALSE)] <- 0
upper1 = reverse1[,ncol(reverse1):1]
# Add them together
merged = as.matrix(upper upper1)
merged
A B C D E
F 1.00 1.72 1.22 1.79 0
G 1.72 1.00 0.36 0.00 8
H 1.22 0.36 0.00 10.00 9
I 1.79 0.00 10.00 1.00 1
J 0.00 8.00 9.00 1.00 1
It then suggests using heatmap(merged) - however, how would you go about having different colours and scales for the upper and lower part of the matrix?
I am happy with using any package including ggplot2 to make this work.
Thanks in advance!
CodePudding user response:
You can subset the relevant parts of the matrix in the data argument of a layer, and use {ggnewscale} to assign different fill scales to different layers. The trick is to declare a fill scale before adding new_scale_fill(), otherwise the order of operations goes wrong (which usually doesn't matter a lot, but here they do).
You can then tweak every individual scale. In the example below I just tweaked the palettes, but you can also adjust limits, breaks, labels etc.
# Assuming code from question has been executed and we have a 'merged' in memory
library(ggplot2)
library(ggnewscale)
# Wide matrix to long dataframe
# Later, we'll be relying on the notion that the dimnames have been
# converted to factor variables to separate out the upper from the lower
# matrix.
df <- reshape2::melt(merged)
ggplot(df, aes(Var1, Var2))
# The first layer, with its own fill scale
geom_raster(
data = ~ subset(.x, as.numeric(Var1) > as.numeric(Var2)),
aes(fill = value)
)
scale_fill_distiller(palette = "Blues")
# Declare new fill scale for the second layer
new_scale_fill()
geom_raster(
data = ~ subset(.x, as.numeric(Var1) < as.numeric(Var2)),
aes(fill = value)
)
scale_fill_distiller(palette = "Reds")
# I'm not sure what to do with the diagonal. Make it grey?
new_scale_fill()
geom_raster(
data = ~ subset(.x, as.numeric(Var1) == as.numeric(Var2)),
aes(fill = value)
)
scale_fill_distiller(palette = "Greys", guide = "none")
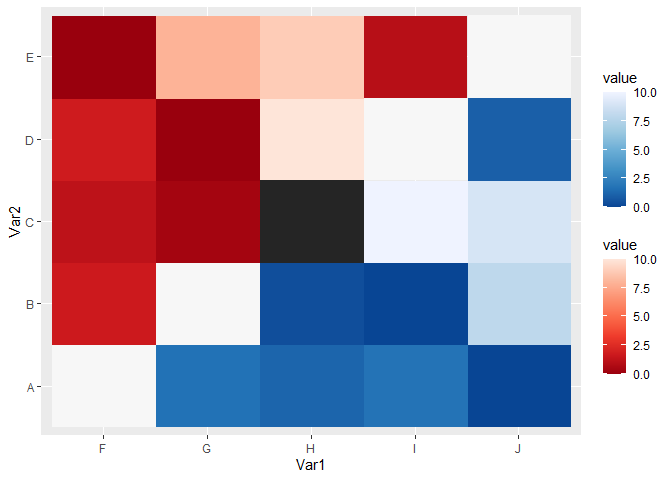
CodePudding user response:
In my opinion @teunbrand's answer is what you're looking for, but another potential option is to use the ComplexHeatmap package, e.g. based on one of the examples in the docs:
library(Matrix)
set.seed(123)
s1<-forceSymmetric(matrix(round(rnorm(25),2),5))
colnames(s1)<-LETTERS[1:5]
rownames(s1)<-LETTERS[6:10]
diag(s1)<-1
s2<-forceSymmetric(matrix(round(rbinom(25,25,0.3),2),5))
colnames(s2)<-LETTERS[1:5]
rownames(s2)<-LETTERS[6:10]
diag(s2)<-1
#Get upper diagonal
reverse = s1[,ncol(s1):1]
diag(reverse) = 0
reverse[lower.tri(reverse, diag = FALSE)] <- 0
upper = reverse[,ncol(reverse):1]
# Get lower diagonal
reverse1 = s2[,ncol(s2):1]
diag(reverse1) = 0
reverse1[upper.tri(reverse1, diag = FALSE)] <- 0
upper1 = reverse1[,ncol(reverse1):1]
# Add them together
m = as.matrix(upper upper1)
m
#> A B C D E
#> F 1.00 1.72 1.22 1.79 0
#> G 1.72 1.00 0.36 0.00 8
#> H 1.22 0.36 0.00 10.00 9
#> I 1.79 0.00 10.00 1.00 1
#> J 0.00 8.00 9.00 1.00 1
library(ComplexHeatmap)
#> Loading required package: grid
#> ========================================
#> ComplexHeatmap version 2.8.0
#> Bioconductor page: http://bioconductor.org/packages/ComplexHeatmap/
#> Github page: https://github.com/jokergoo/ComplexHeatmap
#> Documentation: http://jokergoo.github.io/ComplexHeatmap-reference
#>
#> If you use it in published research, please cite:
#> Gu, Z. Complex heatmaps reveal patterns and correlations in multidimensional
#> genomic data. Bioinformatics 2016.
#>
#> The new InteractiveComplexHeatmap package can directly export static
#> complex heatmaps into an interactive Shiny app with zero effort. Have a try!
#>
#> This message can be suppressed by:
#> suppressPackageStartupMessages(library(ComplexHeatmap))
#> ========================================
library(circlize)
#> ========================================
#> circlize version 0.4.13
#> CRAN page: https://cran.r-project.org/package=circlize
#> Github page: https://github.com/jokergoo/circlize
#> Documentation: https://jokergoo.github.io/circlize_book/book/
#>
#> If you use it in published research, please cite:
#> Gu, Z. circlize implements and enhances circular visualization
#> in R. Bioinformatics 2014.
#>
#> This message can be suppressed by:
#> suppressPackageStartupMessages(library(circlize))
#> ========================================
col1 = colorRamp2(c(-1, 10), c("white", "red"))
col2 = colorRamp2(c(-1, 10), c("white", "blue3"))
# here reordering the symmetric matrix is necessary
od = hclust(dist(m))$order
m = m[od, od]
ht = Heatmap(m, rect_gp = gpar(type = "none"), show_heatmap_legend = FALSE,
cluster_rows = FALSE, cluster_columns = FALSE,
layer_fun = function(j, i, x, y, w, h, fill) {
l = i > j
grid.rect(x[l], y[l], w[l], h[l],
gp = gpar(fill = col1(pindex(m, i[l], j[l])), col = NA))
l = i < j
grid.rect(x[l], y[l], w[l], h[l],
gp = gpar(fill = col2(pindex(m, i[l], j[l])), col = NA))
})
draw(ht, heatmap_legend_list = list(
Legend(title = "Group_A", col_fun = col1),
Legend(title = "Group_B", col_fun = col2)
))
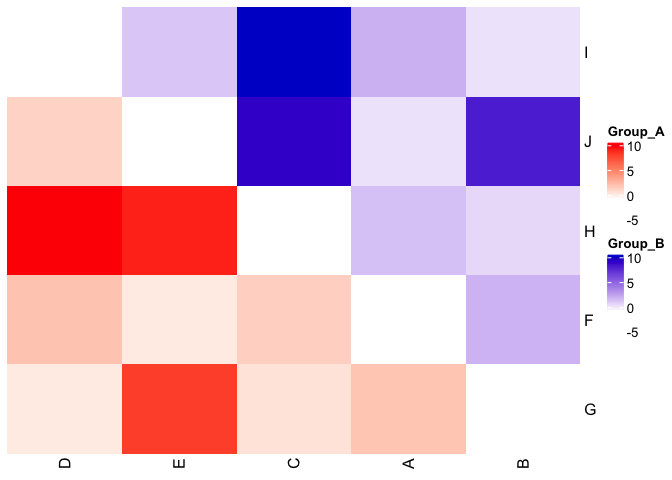
Created on 2022-03-07 by the reprex package (v2.0.1)
