It's my first time creating a code for processing files with a lot of data, so I am kinda stuck here.
What I'm trying to do is to read a list of path, listing all of the csv files that need to be read, retrieve the HEAD and TAIL from each files and put it inside a list.
I have 621 csv files in total, with each files consisted of 5800 rows, and 251 columns
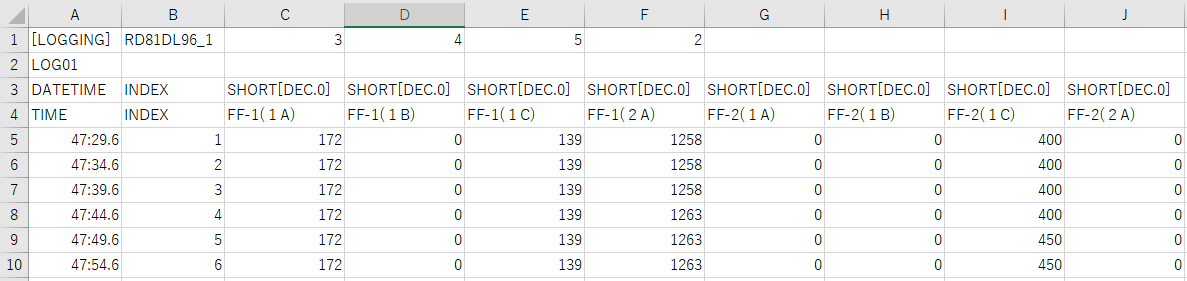 This is the data sample
This is the data sample
[LOGGING],RD81DL96_1,3,4,5,2,,,,
LOG01,,,,,,,,,
DATETIME,INDEX,SHORT[DEC.0],SHORT[DEC.0],SHORT[DEC.0],SHORT[DEC.0],SHORT[DEC.0],SHORT[DEC.0],SHORT[DEC.0],SHORT[DEC.0]
TIME,INDEX,FF-1(1A) ,FF-1(1B) ,FF-1(1C) ,FF-1(2A),FF-2(1A) ,FF-2(1B) ,FF-2(1C),FF-2(2A)
47:29.6,1,172,0,139,1258,0,0,400,0
47:34.6,2,172,0,139,1258,0,0,400,0
47:39.6,3,172,0,139,1258,0,0,400,0
47:44.6,4,172,0,139,1263,0,0,400,0
47:49.6,5,172,0,139,1263,0,0,450,0
47:54.6,6,172,0,139,1263,0,0,450,0
The problem is, while it took about 13 seconds to read all the files (still kinda slow honestly)
But when I add a single line of append code, the process took a lot of times to finish, about 4 minutes.
Below is the snipset of the code:
# CsvList: [File Path, Change Date, File size, File Name]
for x, file in enumerate(CsvList):
timeColumn = ['TIME']
df = dd.read_csv(file[0], sep =',', skiprows = 3, encoding= 'CP932', engine='python', usecols=timeColumn)
# The process became long when this code is added
startEndList.append(list(df.head(1)) list(df.tail(1)))
Why that happened? I'm using dask.dataframe
CodePudding user response:
A first approach using only Python as starting point:
import pandas as pd
import io
def read_first_and_last_lines(filename):
with open(filename, 'rb') as fp:
# skip first 4 rows (headers)
[next(fp) for _ in range(4)]
# first line
first_line = fp.readline()
# start at -2x length of first line from the end of file
fp.seek(-2 * len(first_line), 2)
# last line
last_line = fp.readlines()[-1]
return first_line last_line
data = []
for filename in pathlib.Path('data').glob('*.csv'):
data.append(read_first_and_last_lines(filename))
buf = io.BytesIO()
buf.writelines(data)
buf.seek(0)
df = pd.read_csv(buf, header=None, encoding='CP932')
CodePudding user response:
Currently, your code isn't really leveraging Dask's parallelizing capabilities because:
df.headanddf.tailcalls will trigger a "compute" (i.e., convert your Dask DataFrame into a pandas DataFrame -- which is what we try to minimize in lazy evaluations with Dask), and- the for-loop is running sequentially because you're creating Dask DataFrames and converting them to pandas DataFrames, all inside the loop.
So, your current example is similar to just using pandas within the for-loop, but with the added Dask-to-pandas-conversion overhead.
Since you need to work on each of your files, I'd suggest checking out 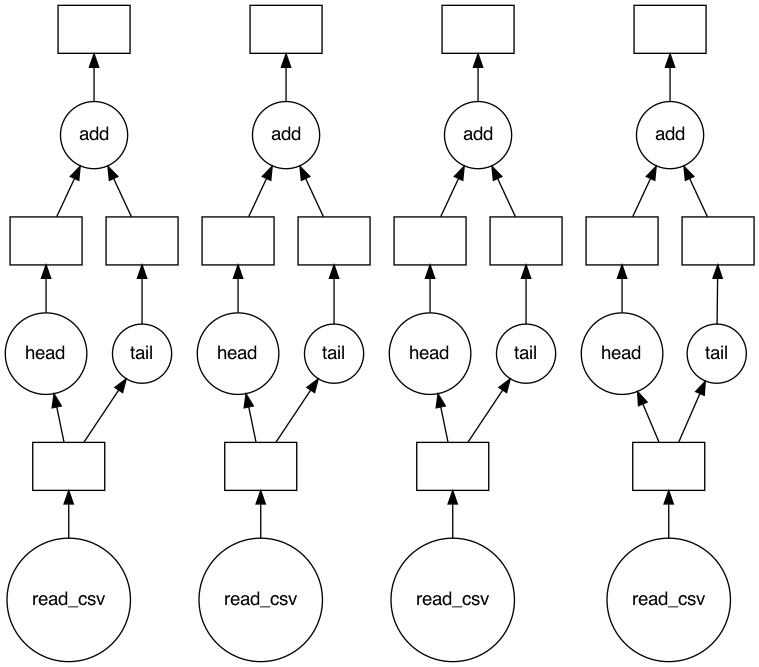
If you really want to use Dask DataFrame here, you may try to:
- read all files into a single Dask DataFrame,
- make sure each Dask "partition" corresponds to one file,
- use Dask Dataframe
applyto get theheadandtailvalues and append them to a new list - call compute on the new list
