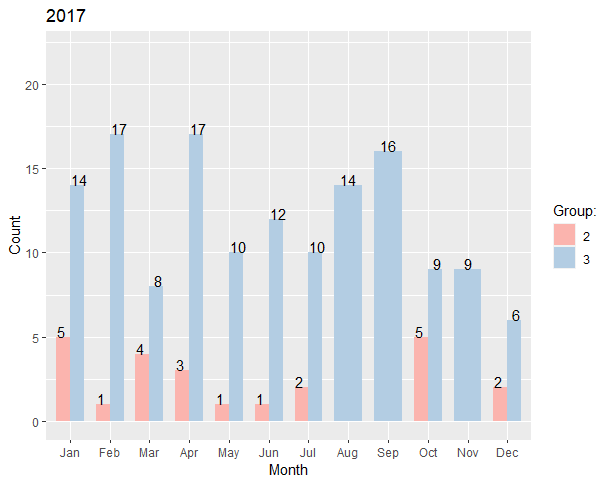
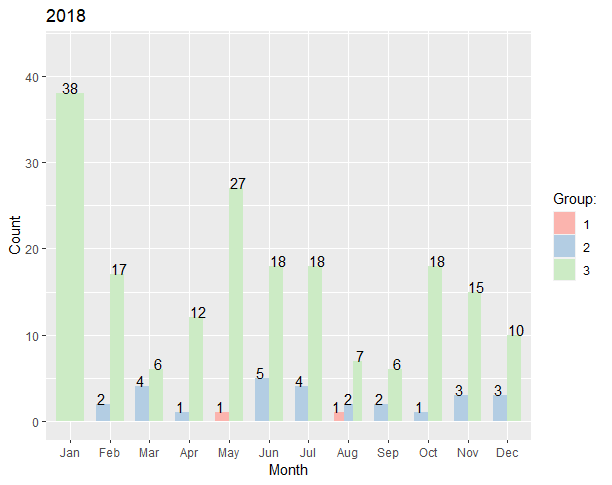
I am creating two sets of grouped bar plots with similar data (one for 2017 and one for 2018). When I create these plots I see that one group does not appear in 2017 (Group 1), but appears in 2018. This is fine since there is no data for this group in 2017. However, I cannot figure out a way to force the legend to recognize this missing group. Also, I see that since this group is missing in 2017, the color scheme shifts. I would like the legends to be uniform regardless if a specific group exists in the data or not. So in this case, I would like the legend to display Groups 1, 2, and 3 even if those aren't in both datasets. Also, I noticed that when a group is missing, the bars get thicker to accommodate for the extra space. Is there a way to keep the bar widths uniform even if some groups are missing?
Here is the code to construct the datasets as well as create the plots I've made so far:
library(dplyr)
library(ggplot2)
#### 2017 Data ###
Year <- rep(2017, 21)
Month <- c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Oct", "Dec", "Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec")
Group <- c(rep(2,9), rep(3,12))
Count <- c(5,1,4,3,1,1,2,5,2,14,17,8,17,10,12,10,14,16,9,9,6)
dta <- as.data.frame(cbind(Year, Month, Group, Count))
dta$Year <- as.numeric(dta$Year)
dta$Month <- as.factor(dta$Month)
dta$Group <- as.factor(dta$Group)
dta$Count <- as.numeric(dta$Count)
tmp <- dta %>%
mutate(Month = factor(Month, levels = c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec")))
### 2017 Plot ###
ggplot(tmp, aes(x = Month, y = Count, fill = Group))
geom_bar(stat = "identity", width = .7, position = position_dodge())
geom_text(aes(label = Count), position = position_dodge(.9), vjust = "bottom")
ggtitle("2017")
guides(fill = guide_legend(title = "Group:"))
scale_fill_brewer(palette = "Pastel1")
ylim(0, max(tmp$Count) 5)
### 2018 Data ###
Year <- rep(2020, 24)
Month <- c("May", "Aug", "Feb", "Mar", "Apr", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec", "Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec")
Group <- c(rep(1,2), rep(2, 10), rep(3, 12))
Count <- c(1,1,2,4,1,5,4,2,2,1,3,3,38,17,6,12,27,18,18,7,6,18,15,10)
dta <- as.data.frame(cbind(Year, Month, Group, Count))
dta$Year <- as.numeric(dta$Year)
dta$Month <- as.factor(dta$Month)
dta$Group <- as.factor(dta$Group)
dta$Count <- as.numeric(dta$Count)
tmp <- dta %>%
mutate(Month = factor(Month, levels = c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec")))
### 2018 Plot ###
ggplot(tmp, aes(x = Month, y = Count, fill = Group))
geom_bar(stat = "identity", width = .7, position = position_dodge())
geom_text(aes(label = Count), position = position_dodge(.9), vjust = "bottom")
ggtitle("2018")
guides(fill = guide_legend(title = "Group:"))
scale_fill_brewer(palette = "Pastel1")
ylim(0, max(tmp$Count) 5)
CodePudding user response:
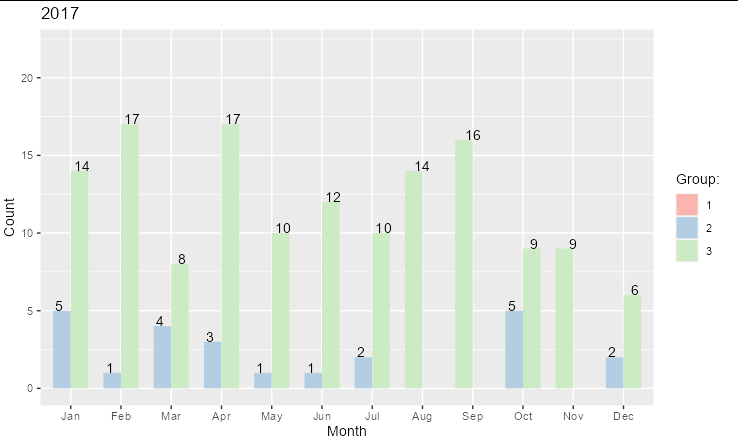
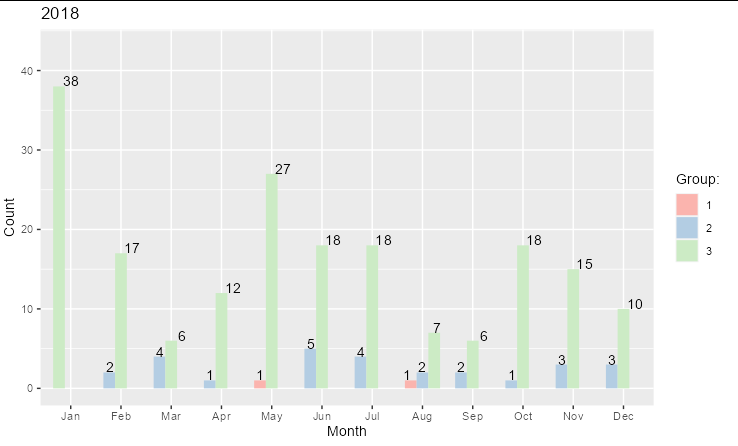
For your 2017 plot, you need to ensure that Group is a factor and contains all the factor levels. You also need to set drop = FALSE in scale_fill_brewer. To prevent the bars from getting wider when a group is missing from a data point, set preserve = "single" inside position_dodge()
ggplot(tmp, aes(x = Month, y = Count,
fill = factor(Group, levels = 1:3)))
geom_bar(stat = "identity", width = .7,
position = position_dodge(preserve = "single"))
geom_text(aes(label = Count),
position = position_dodge(.9), vjust = "bottom")
ggtitle("2017")
guides(fill = guide_legend(title = "Group:"))
scale_fill_brewer(palette = "Pastel1", drop = FALSE)
ylim(0, max(tmp$Count) 5)
You can run the same code with your 2018 set (except changing the ggtitle), and you get: