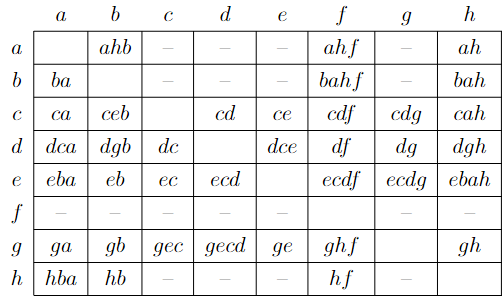
I have an weighted adjacency matrix m and I need the find the shortest path matrix. The expected result is:
library(igraph)
n = 8
m <- t(matrix(c(
0,0,0,0,0,0,0,8,
3,0,0,0,0,0,0,0,
5,0,0,5,1,0,0,0,
0,0,6,0,0,7,1,0,
0,6,2,0,0,0,0,0,
0,0,0,0,0,0,0,0,
7,4,0,0,8,0,0,3,
0,3,0,0,0,9,0,0),ncol=n))
g1 <- graph_from_adjacency_matrix(m, weighted=TRUE, mode="directed")
V(g1)$names <- letters[1:n]
plot(g1, vertex.label = V(g1)$names, edge.arrow.size=0.5)
My attept is:
path_name <- c()
for (i in c(1,2,6,8)){
for (path in all_simple_paths(g1, i, V(g1), mode="out")) {
path_name <- c(path_name, paste(V(g1)[path]$names, collapse=''));
}
}
path_name
[1] "ah" "ahb" "ahf" "ba" "bah" "bahf" "hb" "hba" "hf"
One can see that I have found the paths just for four nodes: 1, 2, 5, 6 with names a, b, f, h. If we take a, b, h as a start node we can obtain 3 paths for each node while for h no path.
Question. How to reconstruct the paths for all nodes? I don't found the Lee (wave) algoritms.
CodePudding user response:
You can use igraph::all_shortest_paths but, unless I am missing something, reconstructing the matrix is still tricky.
EDIT: cleaned up purrr section
library(purrr)
library(igraph)
n = 8
m <- t(matrix(c(
0,0,0,0,0,0,0,8,
3,0,0,0,0,0,0,0,
5,0,0,5,1,0,0,0,
0,0,6,0,0,7,1,0,
0,6,2,0,0,0,0,0,
0,0,0,0,0,0,0,0,
7,4,0,0,8,0,0,3,
0,3,0,0,0,9,0,0),ncol=n))
g1 <- graph_from_adjacency_matrix(m, weighted=TRUE, mode="directed")
V(g1)$names <- letters[1:n]
v <- 1:n
mat <- matrix("",
nrow = n,
ncol = n,
dimnames = list(V(g1)$names, V(g1)$names))
map(v, ~all_shortest_paths(g1, .x, v[-.x], mode = "out")) |>
map(chuck, "res") |>
compact() |>
map_depth(~{V(g1)$names[as.vector(.x)]}, .depth = 2) |>
flatten() |>
walk(~{
mat[first(.x), last(.x)] <<- paste(.x, collapse = "")
})
mat
##> a b c d e f g h
##> a "" "ahb" "" "" "" "ahf" "" "ah"
##> b "ba" "" "" "" "" "bahf" "" "bah"
##> c "ca" "ceb" "" "cd" "ce" "cdf" "cdg" "cdgh"
##> d "dgba" "dgb" "dc" "" "dce" "df" "dg" "dgh"
##> e "eca" "eb" "ec" "ecd" "" "ecdf" "ecdg" "ecdgh"
##> f "" "" "" "" "" "" "" ""
##> g "gba" "gb" "gec" "gecd" "ge" "ghf" "" "gh"
##> h "hba" "hb" "" "" "" "hf" "" ""
CodePudding user response:
You can try the code below
m <- d <- distances(g1, mode = "out")
for (i in row.names(d)) {
for (j in colnames(d)) {
if (i != j) {
if (!is.infinite(d[i, j])) {
m[i, j] <- paste0(names(shortest_paths(g1, i, j)$vpath[[1]]), collapse = "")
} else {
m[i,j] <- NA
}
} else {
m[i, j] <- ""
}
}
}
and you will see the shortest-path matrix m
> m
a b c d e f g h
a "" "ahb" NA NA NA "ahf" NA "ah"
b "ba" "" NA NA NA "bahf" NA "bah"
c "ca" "ceb" "" "cd" "ce" "cdf" "cdg" "cdgh"
d "dga" "dgb" "dc" "" "dce" "df" "dg" "dgh"
e "eca" "eb" "ec" "ecd" "" "ecdf" "ecdg" "ecdgh"
f NA NA NA NA NA "" NA NA
g "ga" "gb" "gec" "gecd" "ge" "ghf" "" "gh"
h "hba" "hb" NA NA NA "hf" NA ""
