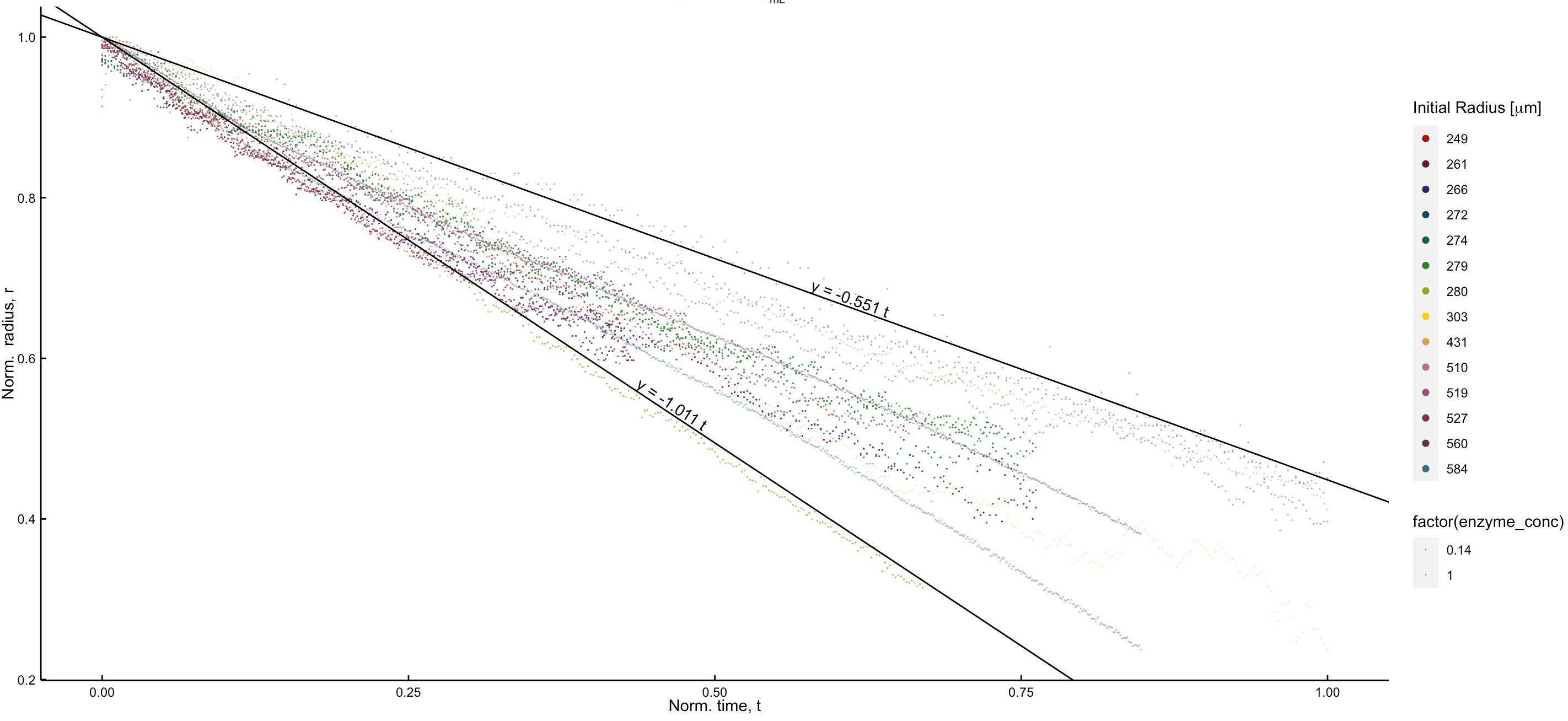
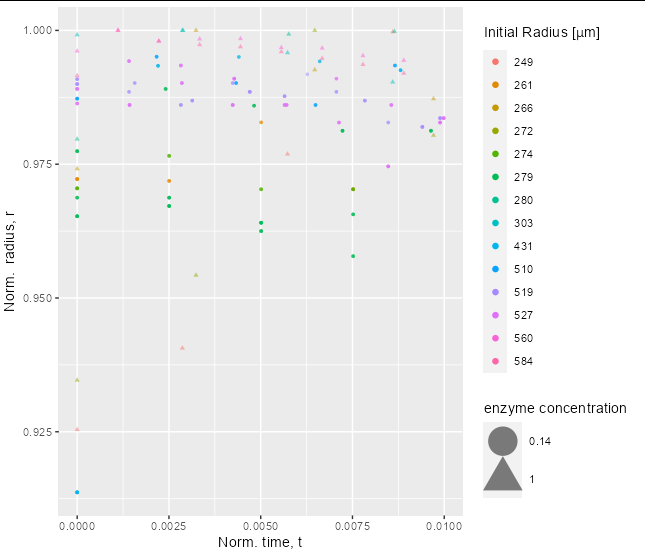
In my figure 2 legends, one for the color and one for the shape. I can use guide to overwrite the aes to make the colors in the legend bigger but I do not know how to overwrite the legend of the shape; see the figure.
Question:
How to use guides to overwrite the legend of the shapes to make the entries there bigger, as I have done for the color legend.
Here is the code I'm using
ggplot(DT, aes(x=norm_time, y=norm_radii))
#facet_wrap(Y~., scales = "free")
geom_point(aes(color = factor(tag), shape = factor(enzyme_conc)), alpha = 0.5, size = 0.2)
labs(x = expression("Norm. time, t"), y = expression("Norm. radius, r"), color = expression("Initial Radius ["*mu*m*"]"))
guides(colour = guide_legend(override.aes = list(linetype = c(rep("blank", length(unique(DT$tag))))
, shape = c(rep(16, length(unique(DT$tag)))), size = c(rep(2, length(unique(DT$tag)))), alpha = 1)))
Here is the dataset I'm using.
structure(list(norm_time = c(0, 0.00286532951289398, 0.00573065902578797,
0.00859598853868195, 0, 0.00323624595469256, 0.00647249190938511,
0.00970873786407767, 0, 0.00323624595469256, 0.00647249190938511,
0.00970873786407767, 0, 0.00288184438040346, 0.00576368876080692,
0.00864553314121037, 0, 0.00286532951289398, 0.00573065902578797,
0.00859598853868195, 0, 0.00111234705228031, 0.00222469410456062,
0.00333704115684093, 0.00444938820912125, 0.00556173526140156,
0.00667408231368187, 0.00778642936596218, 0.00889877641824249,
0, 0.00111234705228031, 0.00222469410456062, 0.00333704115684093,
0.00444938820912125, 0.00556173526140156, 0.00667408231368187,
0.00778642936596218, 0.00889877641824249, 0, 0, 0, 0, 0, 0.0025062656641604,
0.0025062656641604, 0.0025062656641604, 0.0050125313283208, 0.0050125313283208,
0.0075187969924812, 0.0075187969924812, 0.0075187969924812, 0.0075187969924812,
0, 0, 0, 0, 0, 0.0025062656641604, 0.0025062656641604, 0.0025062656641604,
0.0050125313283208, 0.0050125313283208, 0.0050125313283208, 0.0075187969924812,
0.0075187969924812, 0.0075187969924812, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0.00240963855421687, 0.0025062656641604, 0.0025062656641604,
0.00240963855421687, 0.0025062656641604, 0.0025062656641604,
0.00240963855421687, 0.0025062656641604, 0.0025062656641604,
0.0025062656641604, 0.0025062656641604, 0.0025062656641604, 0.00481927710843373,
0.0050125313283208, 0.0050125313283208, 0.0050125313283208, 0.0050125313283208,
0.00481927710843373, 0.0050125313283208, 0.0050125313283208,
0.0050125313283208, 0.00481927710843373, 0.0050125313283208,
0.0050125313283208, 0.0050125313283208, 0.0072289156626506, 0.0075187969924812,
0.0075187969924812, 0.0072289156626506, 0.0075187969924812, 0.0075187969924812,
0.0075187969924812, 0.0075187969924812, 0.0072289156626506, 0.0072289156626506,
0.00963855421686747, 0.00963855421686747, 0.00963855421686747,
0.00963855421686747, 0, 0, 0, 0, 0, 0, 0, 0.00220264317180617,
0.00220264317180617, 0.00220264317180617, 0.00440528634361233,
0.00440528634361233, 0.0066079295154185, 0.0066079295154185,
0.00881057268722467, 0.00881057268722467, 0.00881057268722467,
0, 0, 0, 0, 0.00216450216450216, 0.00216450216450216, 0.00216450216450216,
0.00432900432900433, 0.00432900432900433, 0.00432900432900433,
0.00649350649350649, 0.00649350649350649, 0.00649350649350649,
0.00865800865800866, 0.00865800865800866, 0.00865800865800866,
0, 0, 0, 0, 0, 0, 0, 0.00156739811912226, 0.00141242937853107,
0.00156739811912226, 0.00141242937853107, 0.00313479623824451,
0.00282485875706215, 0.00313479623824451, 0.00282485875706215,
0.00313479623824451, 0.00282485875706215, 0.00470219435736677,
0.00423728813559322, 0.00470219435736677, 0.00470219435736677,
0.00470219435736677, 0.00423728813559322, 0.00626959247648903,
0.00564971751412429, 0.00564971751412429, 0.00564971751412429,
0.00783699059561128, 0.00706214689265537, 0.00706214689265537,
0.00783699059561128, 0.00783699059561128, 0.00940438871473354,
0.00847457627118644, 0.00940438871473354, 0.00940438871473354,
0.00847457627118644, 0.00940438871473354, 0.00988700564971751,
0.00988700564971751, 0.00988700564971751, 0.00988700564971751,
0, 0, 0, 0, 0, 0, 0, 0.00141242937853107, 0.0014265335235378,
0.00141242937853107, 0.00141242937853107, 0.0014265335235378,
0.0014265335235378, 0.0014265335235378, 0.00282485875706215,
0.00285306704707561, 0.00282485875706215, 0.00285306704707561,
0.00282485875706215, 0.00285306704707561, 0.00423728813559322,
0.00427960057061341, 0.00423728813559322, 0.00423728813559322,
0.00427960057061341, 0.00423728813559322, 0.00427960057061341,
0.00427960057061341, 0.00564971751412429, 0.00570613409415121,
0.00564971751412429, 0.00570613409415121, 0.00570613409415121,
0.00564971751412429, 0.00706214689265537, 0.00713266761768902,
0.00706214689265537, 0.00713266761768902, 0.00713266761768902,
0.00847457627118644, 0.00855920114122682, 0.00855920114122682,
0.00847457627118644, 0.00847457627118644, 0.00855920114122682,
0.00847457627118644, 0.00988700564971751, 0.00998573466476462,
0.00988700564971751, 0.00998573466476462, 0.00988700564971751,
0.00998573466476462, 0.00988700564971751, 0.00998573466476462
), norm_radii = c(0.925414688625692, 0.940629276457537, 0.976889499999864,
0.999732576598233, 0.974135688727245, 1, 0.992635601808811, 0.987221126778968,
0.934640522875817, 0.954248366013072, 1, 0.980392156862745, 0.979686761963915,
1, 0.999296213131167, 0.999822382368898, 0.99917181903654, 1,
0.995823611987307, 0.990346548554237, 0.996139131504159, 1, 0.997987488713165,
0.998387853482455, 0.998448504631566, 0.996785224651146, 0.996691603473212,
0.995278275578725, 0.994400786400353, 0.991549345840616, 1, 0.998006223174683,
0.997320331119248, 0.996931508623026, 0.996004525565797, 0.994758729343126,
0.993623516619896, 0.991990575643147, 0.972222222222222, 0.972222222222222,
0.972222222222222, 0.972222222222222, 0.972222222222222, 0.971875,
0.971875, 0.971875, 0.9828125, 0.9828125, 0.9703125, 0.9703125,
0.9703125, 0.9703125, 0.970486111111111, 0.970486111111111, 0.970486111111111,
0.970486111111111, 0.970486111111111, 0.9765625, 0.9765625, 0.9765625,
0.9703125, 0.9703125, 0.9703125, 0.9703125, 0.9703125, 0.9703125,
0.977430555555556, 0.96875, 0.965277777777778, 0.977430555555556,
0.96875, 0.965277777777778, 0.965277777777778, 0.977430555555556,
0.96875, 0.977430555555556, 0.965277777777778, 0.965277777777778,
0.9890625, 0.96875, 0.9671875, 0.9890625, 0.96875, 0.9671875,
0.9890625, 0.96875, 0.9671875, 0.96875, 0.9671875, 0.9671875,
0.9859375, 0.9640625, 0.9625, 0.9640625, 0.9625, 0.9859375, 0.9640625,
0.9640625, 0.9625, 0.9859375, 0.9640625, 0.9640625, 0.9625, 0.98125,
0.9578125, 0.965625, 0.98125, 0.9578125, 0.965625, 0.9578125,
0.965625, 0.98125, 0.98125, 0.98125, 0.98125, 0.98125, 0.98125,
0.913682277318641, 0.913682277318641, 0.913682277318641, 0.913682277318641,
0.913682277318641, 0.913682277318641, 0.913682277318641, 0.993388429752066,
0.993388429752066, 0.993388429752066, 0.99504132231405, 0.99504132231405,
0.994214876033058, 0.994214876033058, 0.992561983471074, 0.992561983471074,
0.992561983471074, 0.987249544626594, 0.987249544626594, 0.987249544626594,
0.987249544626594, 0.995081967213115, 0.995081967213115, 0.995081967213115,
0.99016393442623, 0.99016393442623, 0.99016393442623, 0.986065573770492,
0.986065573770492, 0.986065573770492, 0.99344262295082, 0.99344262295082,
0.99344262295082, 0.990892531876138, 0.989981785063752, 0.990892531876138,
0.989981785063752, 0.990892531876138, 0.989981785063752, 0.989981785063752,
0.99016393442623, 0.988524590163934, 0.99016393442623, 0.988524590163934,
0.986885245901639, 0.986065573770492, 0.986885245901639, 0.986065573770492,
0.986885245901639, 0.986065573770492, 0.988524590163934, 0.99016393442623,
0.988524590163934, 0.988524590163934, 0.988524590163934, 0.99016393442623,
0.991803278688525, 0.987704918032787, 0.987704918032787, 0.987704918032787,
0.986885245901639, 0.988524590163934, 0.988524590163934, 0.986885245901639,
0.986885245901639, 0.981967213114754, 0.982786885245902, 0.981967213114754,
0.981967213114754, 0.982786885245902, 0.981967213114754, 0.983606557377049,
0.983606557377049, 0.983606557377049, 0.983606557377049, 0.989071038251366,
0.986338797814208, 0.989071038251366, 0.986338797814208, 0.986338797814208,
0.989071038251366, 0.989071038251366, 0.994262295081967, 0.986065573770492,
0.994262295081967, 0.994262295081967, 0.986065573770492, 0.986065573770492,
0.986065573770492, 0.99344262295082, 0.99016393442623, 0.99344262295082,
0.99016393442623, 0.99344262295082, 0.99016393442623, 0.986065573770492,
0.990983606557377, 0.986065573770492, 0.986065573770492, 0.990983606557377,
0.986065573770492, 0.990983606557377, 0.990983606557377, 0.986065573770492,
0.986065573770492, 0.986065573770492, 0.986065573770492, 0.986065573770492,
0.986065573770492, 0.990983606557377, 0.982786885245902, 0.990983606557377,
0.982786885245902, 0.982786885245902, 0.974590163934426, 0.986065573770492,
0.986065573770492, 0.974590163934426, 0.974590163934426, 0.986065573770492,
0.974590163934426, 0.982786885245902, 0.983606557377049, 0.982786885245902,
0.983606557377049, 0.982786885245902, 0.983606557377049, 0.982786885245902,
0.983606557377049), enzyme_conc = c(1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14, 0.14,
0.14, 0.14, 0.14), tag = c(249L, 249L, 249L, 249L, 266L, 266L,
266L, 266L, 272L, 272L, 272L, 272L, 280L, 280L, 280L, 280L, 303L,
303L, 303L, 303L, 560L, 560L, 560L, 560L, 560L, 560L, 560L, 560L,
560L, 584L, 584L, 584L, 584L, 584L, 584L, 584L, 584L, 584L, 261L,
261L, 261L, 261L, 261L, 261L, 261L, 261L, 261L, 261L, 261L, 261L,
261L, 261L, 274L, 274L, 274L, 274L, 274L, 274L, 274L, 274L, 274L,
274L, 274L, 274L, 274L, 274L, 279L, 279L, 279L, 279L, 279L, 279L,
279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L,
279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L,
279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L,
279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L, 279L,
279L, 431L, 431L, 431L, 431L, 431L, 431L, 431L, 431L, 431L, 431L,
431L, 431L, 431L, 431L, 431L, 431L, 431L, 510L, 510L, 510L, 510L,
510L, 510L, 510L, 510L, 510L, 510L, 510L, 510L, 510L, 510L, 510L,
510L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L,
519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L,
519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L,
519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 519L, 527L,
527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L,
527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L,
527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L,
527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L,
527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L, 527L)), row.names = c(NA,
-246L), class = c("data.table", "data.frame"), .internal.selfref = <pointer: 0x7fde6080d0e0>)
CodePudding user response:
You can do it in exactly the same way as you did for color. Just put a shape entry inside guides:
ggplot(DT, aes(x = norm_time, y = norm_radii, color = factor(tag)))
geom_point(aes(shape = factor(enzyme_conc)), alpha = 0.5, size = 1)
labs(x = expression("Norm. time, t"),
y = expression("Norm. radius, r"),
color = expression("Initial Radius ["*mu*m*"]"),
shape = 'enzyme concentration')
guides(colour = guide_legend(override.aes = list(shape = 16, size = 2, alpha = 1)),
shape = guide_legend(override.aes = list(size = 10)))
Note also that length-1 values are recycled, so you only need to put shape = 16 and size = 2 inside your override.aes call, if you want all the values to be the same.