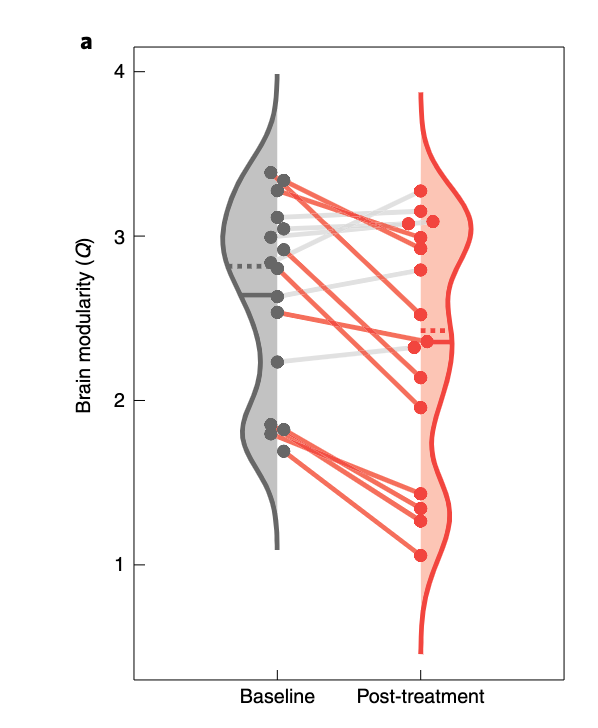
CodePudding user response:
You can get arbitrarily close to a chosen chart using ggplot:
ggplot(df, aes(xval, modularity, color = group))
geom_polygon(data = densdf, aes( x = y, y = x, fill = group), colour = NA)
scale_fill_manual(values = c('#c2c2c2', '#fbc5b4'))
scale_color_manual(values = c('#676767', '#ef453e'))
geom_path(data = densdf, aes(x = y, y = x), size = 2)
geom_segment(color = '#c2c2c2', inherit.aes = FALSE, size = 1.5,
data = df2[df2$`Post-treatment` > df2$Baseline,], alpha = 0.8,
aes(x = 1, xend = 2, y = Baseline, yend = `Post-treatment`))
geom_segment(color = '#ef453e', inherit.aes = FALSE, size = 1.5, alpha = 0.8,
data = df2[df2$`Post-treatment` < df2$Baseline,],
aes(x = 1, xend = 2, y = Baseline, yend = `Post-treatment`))
geom_point(size = 3)
theme_classic()
scale_x_continuous(breaks = 1:2, labels = c('Baseline', 'Post-treatment'),
name = '', expand = c(0.3, 0))
theme(legend.position = 'none',
text = element_text(size = 18, face = 2),
panel.background = element_rect(fill = NA, color = 'black', size = 1.5))
As long as you are prepared to do some work getting your data into the right format:
set.seed(4)
mod <- c(rnorm(16, 2.5, 0.25))
df <- data.frame(modularity = c(mod, mod rnorm(16, -0.25, 0.2)),
xval = rep(c(1, 2), each = 16),
group = rep(c('Baseline', 'Post-treatment'), each = 16),
id = factor(rep(1:16, 2)))
df2 <- df %>% tidyr::pivot_wider(id_cols = id, names_from = group,
values_from = modularity)
BLdens <- as.data.frame(density(df$modularity[1:16])[c('x', 'y')])
PTdens <- as.data.frame(density(df$modularity[17:32])[c('x', 'y')])
BLdens$y <- 1 - 0.25 * BLdens$y
PTdens$y <- 2 0.25 * PTdens$y
densdf <- rbind(BLdens, PTdens)
densdf$group <- rep(c('Baseline', 'Post-treatment'), each = nrow(BLdens))