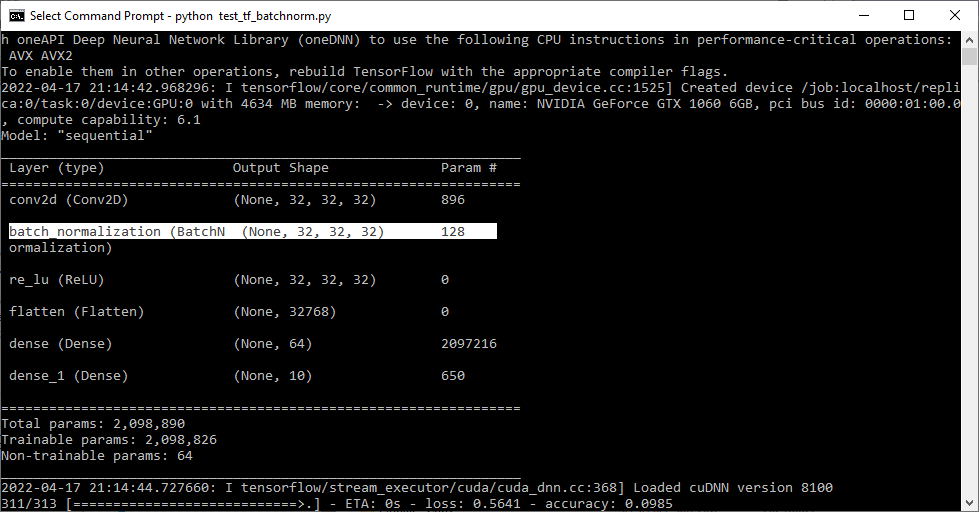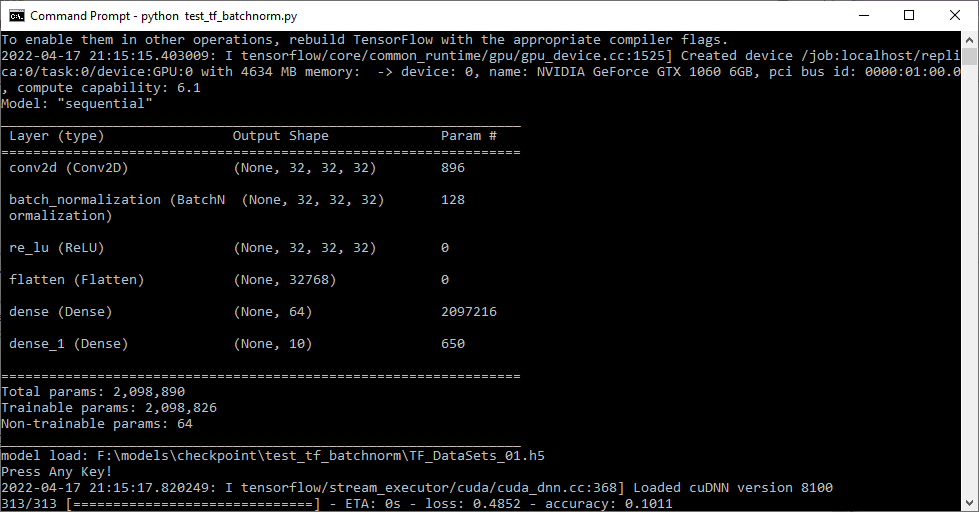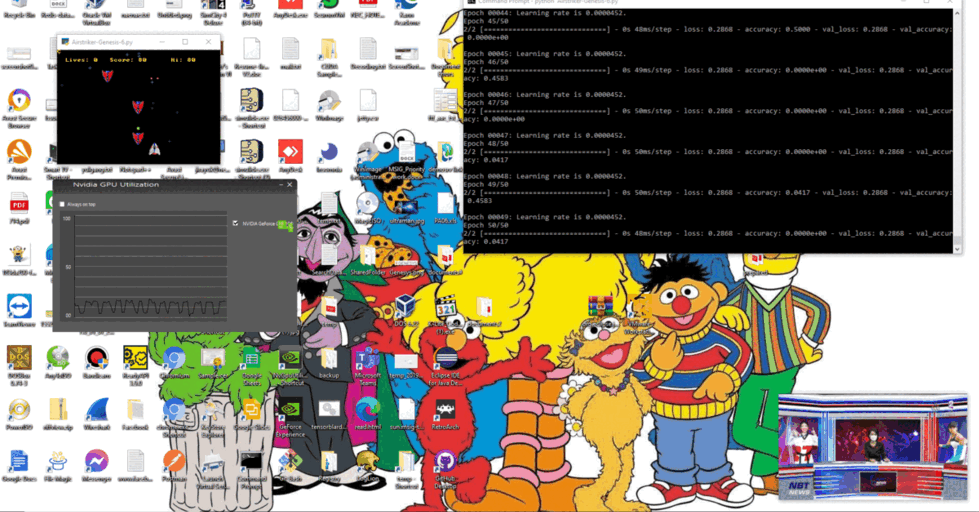
I am trying to save a custom Tensorflow model after 1 epoch training. When the model contains BatchNormalization layer it can not be saved. I can see that "fused_batch_norm" is can not be serialized. How can I call another BatchNormalization layer which can be serialized and saved with both ".h5" and ".pb" format. I am using Tensorflow 2.8 with Tensorflow-metal on MacOS.
def conv_batchnorm_relu(x, filters, kernel_size, strides=1):
# s
x = tf.keras.layers.Conv2D(filters=filters, kernel_size=kernel_size, strides=strides, padding = 'same')(x)
x = tf.keras.layers.BatchNormalization()(x)
x = tf.keras.layers.ReLU()(x)
return x
TypeError: Layer tf.compat.v1.nn.fused_batch_norm was passed non-JSON-serializable arguments. Arguments had types: {'scale': <class 'tensorflow.python.ops.resource_variable_ops.ResourceVariable'>, 'offset': <class 'tensorflow.python.ops.resource_variable_ops.ResourceVariable'>, 'mean': <class 'tensorflow.python.ops.resource_variable_ops.ResourceVariable'>, 'variance': <class 'tensorflow.python.ops.resource_variable_ops.ResourceVariable'>, 'epsilon': <class 'float'>, 'is_training': <class 'bool'>, 'data_format': <class 'str'>}. They cannot be serialized out when saving the model.
CodePudding user response:
You can apply matrix properties or you can create a model with a support function.
Sample:
import os
from os.path import exists
import tensorflow as tf
import h5py
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
Variables
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
filters = 32
kernel_size = (3, 3)
strides = 1
database_buffer = "F:\\models\\buffer\\" os.path.basename(__file__).split('.')[0] "\\TF_DataSets_01.h5"
database_buffer_dir = os.path.dirname(database_buffer)
checkpoint_path = "F:\\models\\checkpoint\\" os.path.basename(__file__).split('.')[0] "\\TF_DataSets_01.h5"
checkpoint_dir = os.path.dirname(checkpoint_path)
loggings = "F:\\models\\checkpoint\\" os.path.basename(__file__).split('.')[0] "\\loggings.log"
if not exists(checkpoint_dir) :
os.mkdir(checkpoint_dir)
print("Create directory: " checkpoint_dir)
if not exists(database_buffer_dir) :
os.mkdir(database_buffer_dir)
print("Create directory: " database_buffer_dir)
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
Functions
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
def conv_batchnorm_relu(filters, kernel_size, strides=1):
model = tf.keras.models.Sequential([
tf.keras.layers.InputLayer(input_shape=( 32, 32, 3 )),
tf.keras.layers.Conv2D(filters=filters, kernel_size=kernel_size, strides=strides, padding = 'same'),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.ReLU(),
])
model.add(tf.keras.layers.Flatten())
model.add(tf.keras.layers.Dense(64))
model.add(tf.keras.layers.Dense(10))
model.summary()
return model
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
DataSet
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
(train_images, train_labels), (test_images, test_labels) = tf.keras.datasets.cifar10.load_data()
# Create hdf5 file
hdf5_file = h5py.File(database_buffer, mode='w')
# Train images
hdf5_file['x_train'] = train_images
hdf5_file['y_train'] = train_labels
# Test images
hdf5_file['x_test'] = test_images
hdf5_file['y_test'] = test_labels
hdf5_file.close()
# Visualize dataset train sample
hdf5_file = h5py.File(database_buffer, mode='r')
x_train = hdf5_file['x_train'][0: 10000]
x_test = hdf5_file['x_test'][0: 100]
y_train = hdf5_file['y_train'][0: 10000]
y_test = hdf5_file['y_test'][0: 100]
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
: Optimizer
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
optimizer = tf.keras.optimizers.Nadam( learning_rate=0.0001, beta_1=0.9, beta_2=0.999, epsilon=1e-07, name='Nadam' )
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
: Loss Fn
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
lossfn = tf.keras.losses.MeanSquaredLogarithmicError(reduction=tf.keras.losses.Reduction.AUTO, name='mean_squared_logarithmic_error')
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
: Model Summary
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
model = conv_batchnorm_relu(filters, kernel_size, strides=1)
model.compile(optimizer=optimizer, loss=lossfn, metrics=['accuracy'])
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
: FileWriter
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
if exists(checkpoint_path) :
model.load_weights(checkpoint_path)
print("model load: " checkpoint_path)
input("Press Any Key!")
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
: Training
"""""""""""""""""""""""""""""""""""""""""""""""""""""""""
history = model.fit(x_train, y_train, epochs=1 ,validation_data=(x_train, y_train))
model.save_weights(checkpoint_path)
input('...')
CodePudding user response:
What I understood from your given template is, you can not use support function this way for adding custom layers in your model. Typically you inherit from keras.Model when you need the model methods like: Model.fit ,Model.evaluate, and Model.save
Here is a example code for better understanding
class ExampleBlock(tf.keras.Model):
def __init__(self, kernel_size, filter1,filter2):
super(ExampleBlock, self).__init__(name='')
self.conv2a = tf.keras.layers.Conv2D(filters1, (1, 1))
self.bn2a = tf.keras.layers.BatchNormalization()
self.conv2b = tf.keras.layers.Conv2D(filters2, kernel_size, padding='same')
self.bn2b = tf.keras.layers.BatchNormalization()
# You can as many layer you like
def call(self, input_tensor, training=False):
x = self.conv2a(input_tensor)
x = self.bn2a(x, training=training)
x = tf.nn.relu(x)
x = self.conv2b(x)
x = self.bn2b(x, training=training)
x = tf.nn.relu(x)
return tf.nn.relu(x)
After defining your model you can create checkpoint after each epoch by defining the call back function.
import keras
model_save_path = "/content/drive/MyDrive/Project/" #path for saving checkpoint
checkpoint = keras.callbacks.ModelCheckpoint(model_save_path 'checkpoint_{epoch:02d}', save_freq='epoch')
You can use this checkpoint in the callback funtion
H = model.fit(
aug.flow(trainX, trainY, batch_size=BS), validation_data=(validX, validY), steps_per_epoch=len(trainX) // BS, epochs=EPOCHS,
class_weight=classWeight,
verbose=1,callbacks=[checkpoint])