I'm trying to apply some equations to get the proportion of a line (numeric array) that coincides with another line (another numeric array). I have a data frame with the required values and I try to create a new column with the percentage outcome based on how the two lines coincide. I have checked the code (below) with some examples and it works OK, but when I apply the case_when() to the data frame, the output is not what it should be. I'll give you a basic example.
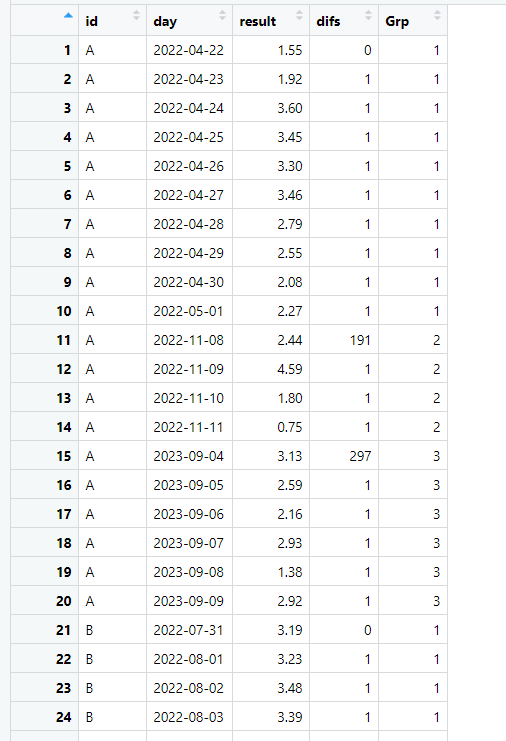
This is my output. It has a 'ID' column [char], a 'date' (days) column [dttm], a 'result' (value) column [double], 'difs' column is the number of days between the previous row [int], and 'Grp' column, that is a subgrouping value.
Here is the code I'm using. The idea is to get the previous value and calculate what % of the vector lies inside another vector whose limits are [2,3]. Right now I'm only checking if the conditions are right for every row. However, when it should get '0', get 'A', or sometimes 'Inf' when it should get 'A', etc, etc. I don't understand why. I think mutate iterate independently over every row inside the group, so I don't understand why the outcomes are wrong, compared to checking by hand.
Rsup = 3 # Highlimit of target array
Rinf = 2 # Low limit of target array
example_output = example%>%
arrange(id,Grp,day) %>%
group_by(id,Grp) %>% # Group by episodes (id Grp)
mutate(from_r = lag(result)) %>% # get previous result y(t-1)
filter(difs != 0, difs < 181) %>% # dischard first sample of every subgroup/episode
mutate(
p_days = case_when(
(min(result,from_r) < Rinf) & (max(result,from_r) > Rsup) ~ 'A',
(min(result,from_r) > Rinf) & (max(result,from_r) < Rsup) ~ '100',
(min(result,from_r) < Rinf) & (max(result,from_r) > Rinf) ~ 'Inf',
(min(result,from_r) < Rsup) & (max(result,from_r) > Rsup) ~ 'Sup',
TRUE ~ '0')
)
# Case 'A': check interval yt - yt-1 cuts target array for both limits
# Case '100': all the interval yt - yt-1 is inside target array (100%)
# Case 'Inf': interval cuts low limit of target array
# Case 'Sup': interval cuts high limit of target array
# Case True ~ '0': interval does not cut target array and it is not inside (0%)
This is how to create the basic example:
structure(list(id = c("A", "A", "A", "A", "A", "A", "A", "A",
"A", "A", "A", "A", "A", "A", "A", "A", "A", "A", "A", "A", "B",
"B", "B", "B", "B", "B", "B", "B", "B", "B", "B", "B", "B", "B",
"B", "B", "B", "B", "B", "B"), day = structure(c(19104, 19105,
19106, 19107, 19108, 19109, 19110, 19111, 19112, 19113, 19304,
19305, 19306, 19307, 19604, 19605, 19606, 19607, 19608, 19609,
19204, 19205, 19206, 19207, 19208, 19209, 19210, 19211, 19212,
19213, 19214, 19215, 19216, 19217, 19218, 19219, 19220, 19221,
19222, 19223), class = "Date"), result = c(1.55, 1.92, 3.6, 3.45,
3.3, 3.46, 2.79, 2.55, 2.08, 2.27, 2.44, 4.59, 1.8, 0.75, 3.13,
2.59, 2.16, 2.93, 1.38, 2.92, 3.19, 3.23, 3.48, 3.39, 2.62, 2.66,
3.77, 3.44, 3.06, 2.59, 2.87, 1.97, 2.5, 2.84, 1.48, 3.04, 2.62,
0.76, 2.74, 2.84), difs = c(0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 191,
1, 1, 1, 297, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1), Grp = c(1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1)), row.names = c(NA, -40L), groups = structure(list(
id = c("A", "B"), .rows = structure(list(1:20, 21:40), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -2L), .drop = TRUE), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"))
Of course, if someone knows a function to get the same output I'm trying with mutate case_when, it will be very helpful too. Thanks in advance.
EDIT: I think mutate iterate independently over every row inside the group, so I don't understand why the outcomes are wrong. Maybe it mixes the results (and from_r) values of every group somehow?
CodePudding user response:
The issue is that you use min/max instead of the vectorized pmin/pmax:
library(dplyr)
ex1 <- example %>%
arrange(id,Grp,day) %>%
group_by(id,Grp) %>% # Group by episodes (id Grp)
mutate(from_r = lag(result)) %>% # get previous result y(t-1)
filter(difs != 0, difs < 181) # dischard first sample of every subgroup/episode
ex1 %>%
mutate(
p_days = case_when(
(pmin(result,from_r) < Rinf) & (pmax(result,from_r) > Rsup) ~ 'A',
(pmin(result,from_r) > Rinf) & (pmax(result,from_r) < Rsup) ~ '100',
(pmin(result,from_r) < Rinf) & (pmax(result,from_r) > Rinf) ~ 'Inf',
(pmin(result,from_r) < Rsup) & (pmax(result,from_r) > Rsup) ~ 'Sup',
TRUE ~ '0')
)
#> # A tibble: 36 × 7
#> # Groups: id, Grp [4]
#> id day result difs Grp from_r p_days
#> <chr> <date> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 A 2022-04-23 1.92 1 1 1.55 0
#> 2 A 2022-04-24 3.6 1 1 1.92 A
#> 3 A 2022-04-25 3.45 1 1 3.6 0
#> 4 A 2022-04-26 3.3 1 1 3.45 0
#> 5 A 2022-04-27 3.46 1 1 3.3 0
#> 6 A 2022-04-28 2.79 1 1 3.46 Sup
#> 7 A 2022-04-29 2.55 1 1 2.79 100
#> 8 A 2022-04-30 2.08 1 1 2.55 100
#> 9 A 2022-05-01 2.27 1 1 2.08 100
#> 10 A 2022-11-09 4.59 1 2 2.44 Sup
#> # … with 26 more rows
To see the difference more clearly check:
min(ex1$result,ex1$from_r) < Rinf
#> [1] TRUE
pmin(ex1$result,ex1$from_r) < Rinf
#> [1] TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE
#> [13] FALSE FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [25] FALSE FALSE FALSE TRUE TRUE FALSE TRUE TRUE FALSE TRUE TRUE FALSE
As you see, using min the condition min(ex1$result,ex1$from_r) < Rinf gives TRUE for each row of your data. If you want to check conditions or set per row you have to use pmin/pmax.