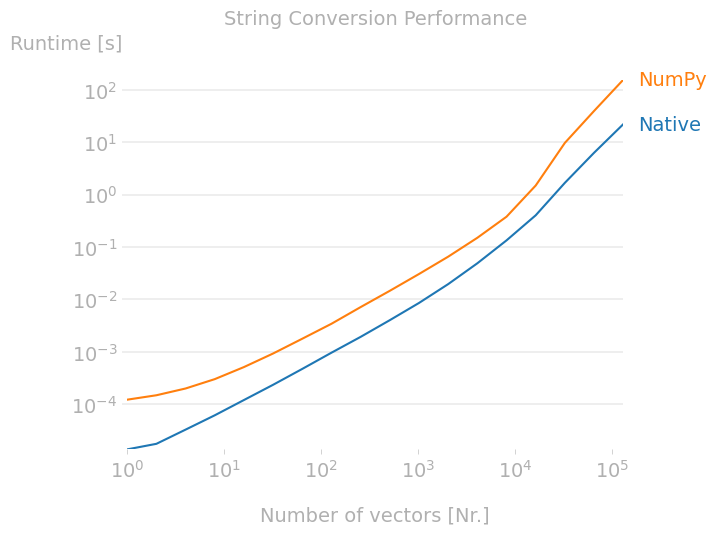
I'm using NumPy's array2string function to convert the values in arrays into a string format for writing to a ascii file. It's simple and relatively quick for large arrays, and out performs a native python operating of string formatting in loop or with map.
aa = np.array2string(array.flatten(), precision=precision, separator=' ', max_line_width=(precision 4) * ncolumns, prefix=' ', floatmode='fixed')
aa = ' ' aa[1:-1] '\n'
However, when testing with small arrays I've noticed some strange results when the number of elements in the array is less than a few thousand. I've run a quick comparison with a native python approach using map and join and performance-wise, it does what I expect - gets much slower as the array gets quite large and it's quicker for very small arrays because of the overhead of the numpy function.
I've used the perfplot to run a benchmark and show what I mean: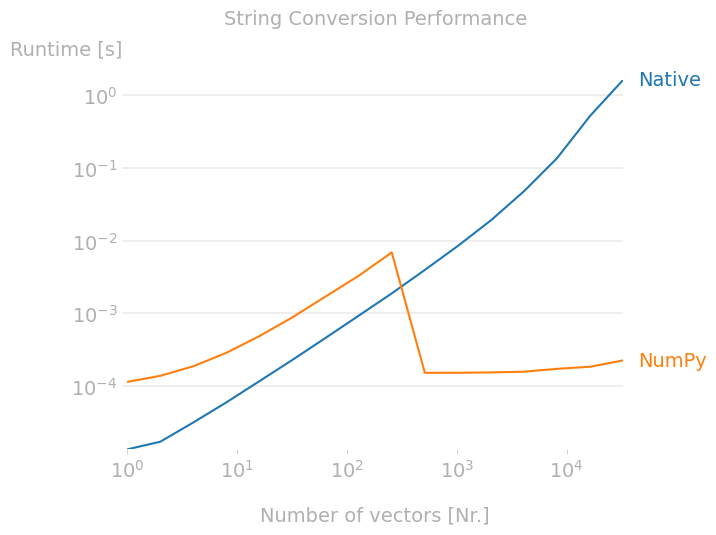
Does anyone know what is the cause of the strange spike in the numpy.array2string method from above? It's actually slower for a (100, 3) array than a (500000,3) array.
I'm just curious as to what is happening, the numpy solution is still the best option for the likely size of my data (>1000) but the spike seems weird.
Update - Full code added
Here's the full script I'm running on my computer:
import numpy as np
import perfplot
precision = 16
ncolumns = 6
# numpy method
def numpystring(array, precision, ncolumns):
indent = ' '
aa = np.array2string(array.flatten(), precision=precision, separator=' ', max_line_width=(precision 6) * ncolumns,
prefix=' ', floatmode='fixed')
return indent aa[1:-1] '\n'
# native python string creation
def nativepython_string(array, precision, ncolumns):
fmt = '{' f":.{precision}f" '}'
data_str = ''
# calculate number of full rows
if array.size <= ncolumns:
nrows = 1
else:
nrows = int(array.size / ncolumns)
# write full rows
for row in range(nrows):
shift = row * ncolumns
data_str = ' ' ' '.join(
map(lambda x: fmt.format(x), array.flatten()[0 shift:ncolumns shift])) '\n'
# write any remaining data in last non-full row
if array.size > ncolumns and array.size % ncolumns != 0:
data_str = ' ' ' '.join(
map(lambda x: fmt.format(x), array.flatten()[ncolumns shift::])) '\n'
return data_str
# Benchmark methods
out = perfplot.bench(
setup=lambda n: np.random.random([n,3]), # setup random nx3 array
kernels=[
lambda a: nativepython_string(a, precision, ncolumns),
lambda a: numpystring(a, precision, ncolumns)
],
equality_check=None,
labels=["Native", "NumPy"],
n_range=[2**k for k in range(16)],
xlabel="Number of vectors [Nr.]",
title="String Conversion Performance"
)
out.show(
time_unit="us", # set to one of ("auto", "s", "ms", "us", or "ns") to force plot units
)
out.save("perf.png", transparent=True, bbox_inches="tight")
Hope this helps.
Update 2 - Fixed benchmark function
Apparently the threshold parameter was the cause. Here's the fixed function:
# numpy method
def numpystring(array, precision, ncolumns):
indent = ' '
aa = np.array2string(array.flatten(), precision=precision, separator=' ', max_line_width=(precision 6) * ncolumns,
prefix=' ', floatmode='fixed', threshold=sys.maxsize)
return indent aa[1:-1] '\n'
Seems the numpy function is actually slower than the native solution.
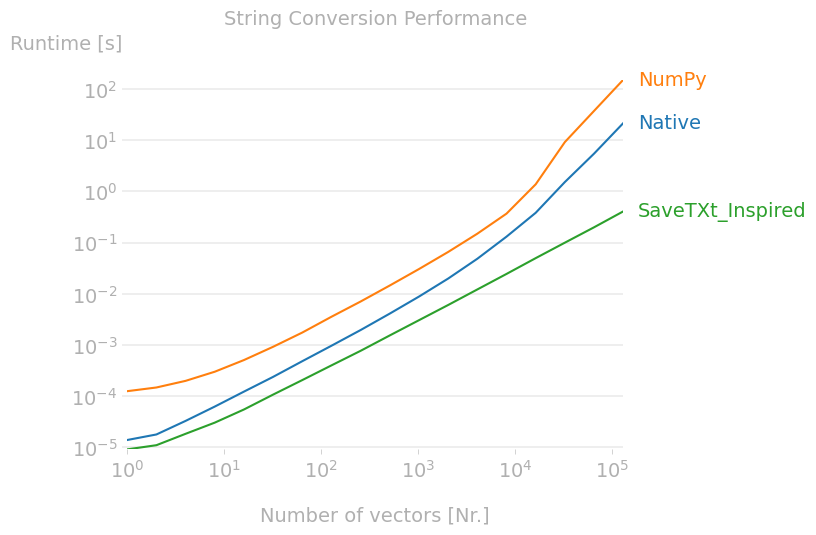
Update 3 - Better solution.
As suggested in the answer below the method used in savetxt may be quicker. A quick implementation shows this to be true.
CodePudding user response:
A sample of using savetxt with small 2d array:
In [87]: np.savetxt('test.txt', np.arange(24).reshape(3,8), fmt=']')
In [88]: cat test.txt
0 1 2 3 4 5 6 7
8 9 10 11 12 13 14 15
16 17 18 19 20 21 22 23
In [90]: np.savetxt('test.txt', np.arange(24).reshape(3,8), fmt=']', newline=' ')
In [91]: cat test.txt
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23
It constructs a fmt string, based on the parameter and number of columns:
In [95]: fmt=' '.join([']']*8)
In [96]: fmt
Out[96]: '] ] ] ] ] ] ] ]'
and then writes this line to the file:
In [97]: fmt%tuple(np.arange(8))
Out[97]: ' 0 1 2 3 4 5 6 7'