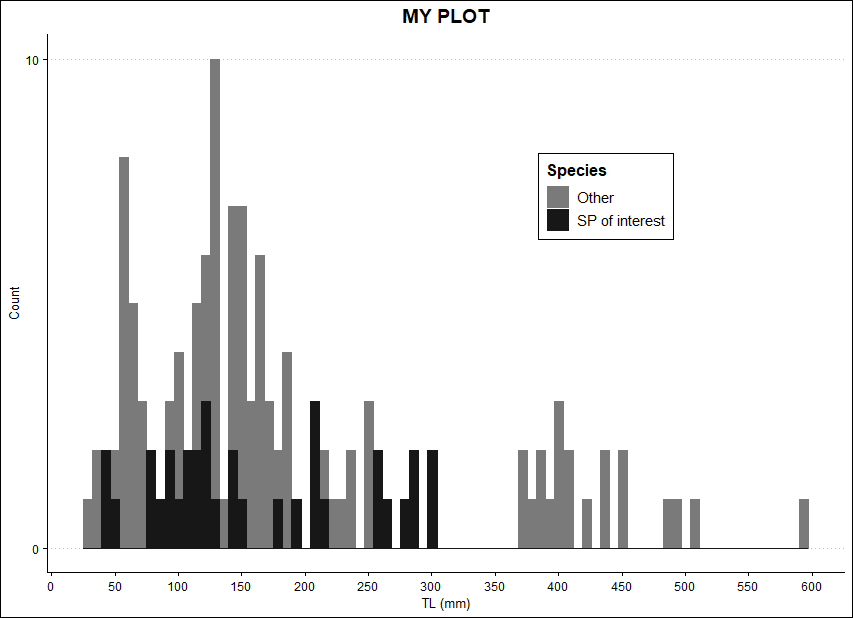
I am trying to plot a second variable on top an existing histogram in ggplot and I am not sure how to achieve this or what this is called. Basically, similar to adding a density line but with data from another column (different variable than the one used for the histogram). Here is an example:
My data (only 3% of my actual data for privacy purposes and to keep the code short):
structure(list(mm = c(88L, 57L, 260L, 100L, 401L, 67L, 146L, 162L, 406L, 60L, 150L, 237L, 154L, 425L, 142L, 65L, 180L, 35L, 147L, 50L, 126L, 32L, 45L, 403L, 144L, 170L, 288L, 150L, 249L, 146L, 174L, 433L, 126L, 85L, 136L, 55L, 132L, 132L, 97L, 388L, 120L, 130L, 210L, 121L, 597L, 453L, 40L, 101L, 80L, 196L, 215L, 224L, 185L, 164L, 39L, 195L, 249L, 165L, 209L, 49L, 510L, 143L, 180L, 131L, 390L, 96L, 436L, 166L, 69L, 162L, 64L, 111L, 119L, 131L, 213L, 64L, 54L, 178L, 48L, 57L, 265L, 132L, 485L, 147L, 103L, 70L, 60L, 284L, 157L, 112L, 256L, 187L, 141L, 151L, 215L, 78L, 375L, 107L, 94L, 235L, 94L, 369L, 118L, 143L, 249L, 448L, 127L, 121L, 383L, 119L, 100L, 162L, 45L, 69L, 187L, 64L, 156L, 204L, 140L, 280L, 120L, 261L, 229L, 122L, 185L, 175L, 130L, 141L, 82L, 58L, 106L, 115L, 151L, 94L, 112L, 60L, 400L, 122L, 113L, 110L, 114L, 148L, 303L, 376L, 406L, 301L, 129L, 122L, 495L, 90L, 152L), YP = c("YP", "other", "YP", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "YP", "other", "other", "other", "other", "other", "other", "YP", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "YP", "other", "other", "other", "YP", "YP", "other", "other", "YP", "other", "YP", "other", "other", "other", "other", "other", "other", "YP", "other", "other", "YP", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "YP", "other", "other", "YP", "other", "other", "YP", "YP", "other", "other", "other", "other", "other", "YP", "other", "other", "other", "other", "YP", "other", "other", "other", "other", "YP", "other", "other", "YP", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "other", "YP", "other", "other", "other", "other", "YP", "other", "YP", "YP", "YP", "other", "other", "other", "other", "other", "YP", "YP", "other", "other", "other", "YP", "YP", "other", "other", "other", "other", "YP", "YP", "YP", "other", "YP", "other", "other", "YP", "other", "YP", "other", "other", "other"), TS = c(72.2364762348789, 56.072368188802,
112.558570306398, 76.9944343224768, 128.685244324351, 62.0886592937318,
91.0798384599843, 94.9503355973493, 129.146464600376, 57.9815046059066,
92.0858456462626, 109.11119845305, 93.0653758618353, 130.848758867367,
90.0458830839189, 60.9606941855156, 98.8718539901336, 37.920054848636,
91.3339008799045, 51.1954962620356, 85.596412576734, 34.5846903022058,
47.2739778692513, 128.870418835656, 90.5664510102187, 96.7444178270107,
116.36538907066, 92.0858456462626, 110.949596806416, 91.0798384599843,
97.6100382367648, 131.542858236253, 85.596412576734, 70.9454797665696,
88.4390148470959, 54.7429411543526, 87.3278875586647, 87.3278875586647,
75.8607426198959, 127.458618740778, 83.7804426663477, 86.7596322459568,
104.609342293304, 84.0893241071107, 143.497154960473, 133.223504910479,
42.8900932821207, 77.3647856368317, 68.6890313425619, 102.04142761656,
105.485147406912, 107.011466050045, 99.8916438094152, 95.4070284436167,
41.9477644689454, 101.851043569743, 110.949596806416, 95.6332905385796,
104.43168085813, 50.4435514956775, 137.634767211238, 90.3070771382737,
98.8718539901336, 87.0448443695468, 127.649981630184, 75.4750396864329,
131.799844297451, 95.8581854826305, 63.18344410111, 94.9503355973493,
60.383628362647, 80.8787140928451, 83.4689764136111, 87.0448443695468,
105.137294407705, 60.383628362647, 54.0599862131223, 98.4559837418714,
49.6761016259917, 56.072368188802, 113.267544123207, 87.3278875586647,
135.764021720693, 91.3339008799045, 78.0946129419071, 63.7189929090771,
57.9815046059066, 115.84482114436, 93.783468875064, 81.2125279896034,
111.981504483529, 100.291862719328, 89.7828431198754, 92.3331555262515,
105.485147406912, 67.7467025293866, 126.190186686619, 79.5126892186721,
74.6914317960895, 108.795772836446, 74.6914317960895, 125.589851091188,
83.1548817226121, 90.3070771382737, 110.949596806416, 132.810404110486,
85.8906433579888, 84.0893241071107, 126.975862297304, 83.4689764136111,
76.9944343224768, 94.9503355973493, 47.2739778692513, 63.18344410111,
100.291862719328, 60.383628362647, 93.5456405898278, 103.530426170882,
89.5179309695183, 115.316869029959, 83.7804426663477, 112.701449560551,
107.833132972289, 84.3956632849718, 99.8916438094152, 97.8233339494332,
86.7596322459568, 89.7828431198754, 69.6080903831755, 56.7196888525377,
79.1632030828511, 82.1963738176802, 92.3331555262515, 74.6914317960895,
81.2125279896034, 57.9815046059066, 128.592310443359, 84.3956632849718,
81.5433746124733, 80.5418792147937, 81.8713062492431, 91.5862408295004,
118.255135021059, 126.289307916972, 129.146464600376, 118.008644053953,
86.4722176903414, 84.3956632849718, 136.523639922807, 73.0729159296925,
92.5788329858984)), class = "data.frame", row.names = c(NA, -151L ))
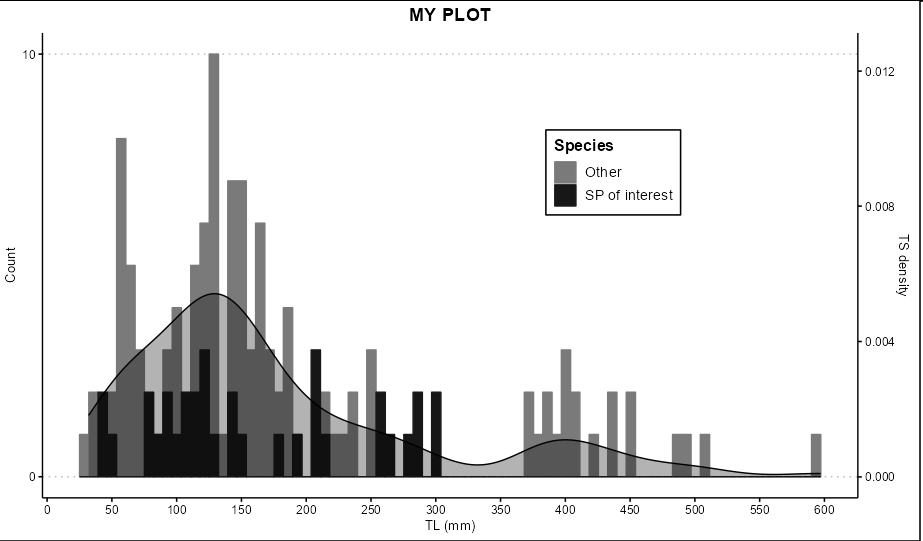
The histogram I have, to which I would like to plot 'TS' on top of it similar to adding density curve I guess but with a second variable (actual histogram with my entire data looks nicer, just an example):
ggplot(DATA, aes(x=mm, color = YP, fill = YP))
geom_histogram(bins = 80, position="identity")
scale_x_continuous(name = "TL (mm)", breaks = seq(0, 800, 50))
scale_y_continuous(name = "Count", breaks = seq(0,290, 10))
scale_fill_manual(name = "Species",
labels = c("Other", "SP of interest"),
values = c("gray48", "gray9"))
scale_color_manual(name = "Species",
labels = c("Other", "SP of interest"),
values = c("gray48", "gray9"))
ggtitle("MY PLOT")
theme_clean()
theme(plot.title = element_text(hjust = 0.5), legend.position = c(0.7, 0.7))
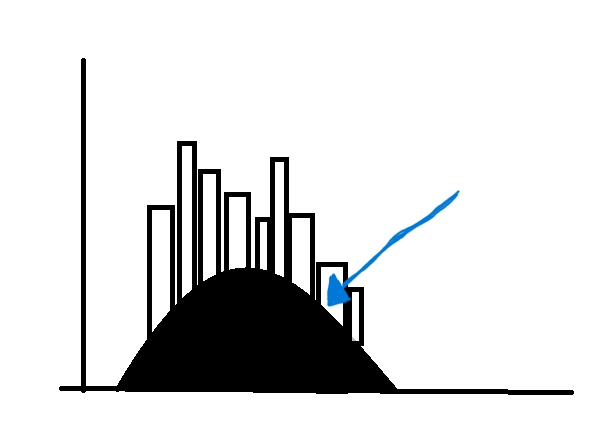
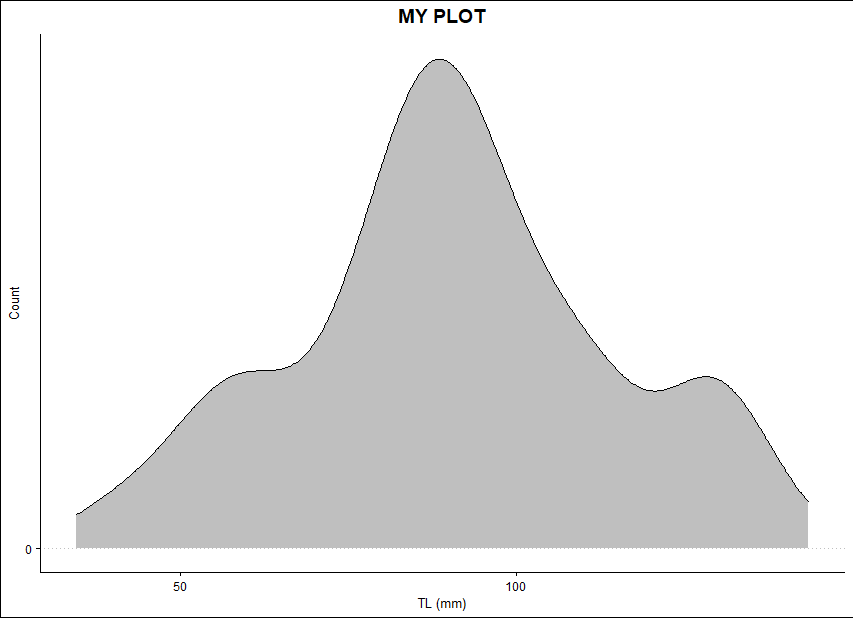
How can I add the 'TS' variable on top of this histogram (similar to a density curve but with the TS variable)? My goal is to end with something like this:
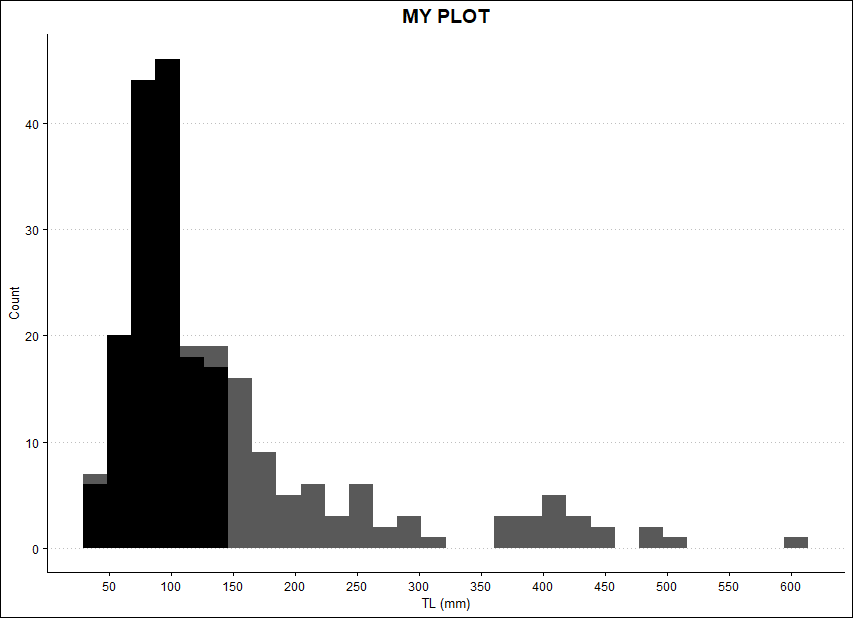
I tried the following, but it just plots another histogram on top, when I want a curve:
ggplot(DATA)
geom_histogram(aes(x=mm))
geom_histogram(aes(x=TS), fill = 'black')
scale_x_continuous(name = "TL (mm)", breaks = seq(0, 700, 50))
scale_y_continuous(name = "Count", breaks = seq(0,50, 10))
ggtitle("MY PLOT")
theme_clean()
theme(plot.title = element_text(hjust = 0.5), legend.position = c(0.7, 0.7))
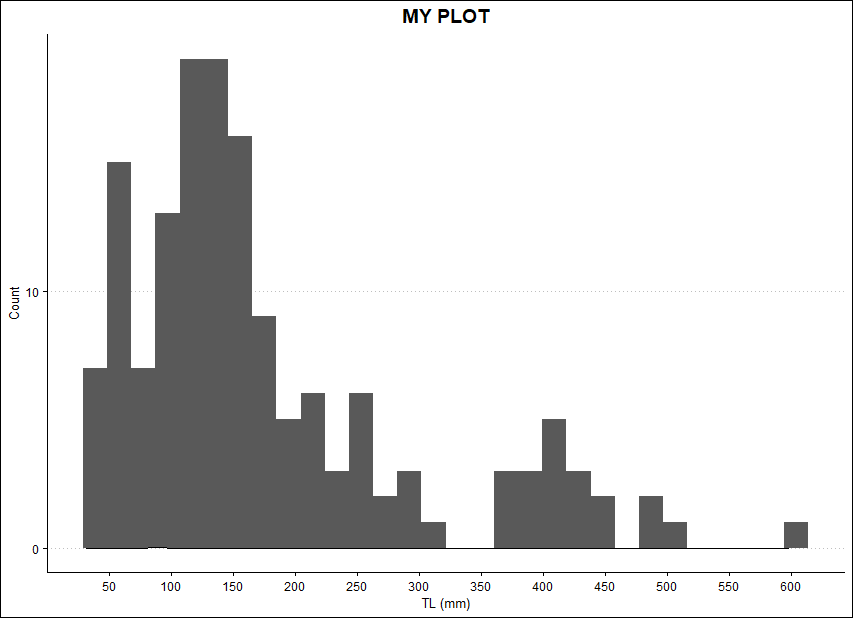
I tried doing something similar with geom_density, but not much happens:
ggplot(DATA)
geom_histogram(aes(x=mm))
geom_density(aes(x=TS), fill = 'black', color='black', alpha = 0.25)
scale_x_continuous(name = "TL (mm)", breaks = seq(0, 700, 50))
scale_y_continuous(name = "Count", breaks = seq(0,50, 10))
ggtitle("MY PLOT")
theme_clean()
theme(plot.title = element_text(hjust = 0.5), legend.position = c(0.7, 0.7))
Maybe something like this but plotting it on top of my histogram...
ggplot(DATA)
geom_density(aes(x=TS), fill = 'black', color='black', alpha = 0.25)
scale_x_continuous(name = "TL (mm)", breaks = seq(0, 700, 50))
scale_y_continuous(name = "Count", breaks = seq(0,50, 10))
ggtitle("MY PLOT")
theme_clean()
theme(plot.title = element_text(hjust = 0.5), legend.position = c(0.7, 0.7))
I have not found examples online to be able to achieve what I want. Thank you!
CodePudding user response:
You can use geom_density, but with the density multiplied large enough to see, and a secondary axis to give an accurate read-out of the density. To get this to work you will need to scale the density to a similar scale to the histogram (with your sample data multiplying by 800 is about right).
In the scale_y_continuous call, add a sec.axis that reverses the transformation:
ggplot(DATA, aes(x=mm, color = YP, fill = YP))
geom_histogram(bins = 80, position="identity")
scale_x_continuous(name = "TL (mm)", breaks = seq(0, 800, 50))
scale_fill_manual(name = "Species",
labels = c("Other", "SP of interest"),
values = c("gray48", "gray9"))
scale_color_manual(name = "Species",
labels = c("Other", "SP of interest"),
values = c("gray48", "gray9"))
ggtitle("MY PLOT")
ggthemes::theme_clean()
theme(plot.title = element_text(hjust = 0.5), legend.position = c(0.7, 0.7))
geom_density(aes(y = ..density.. * 800), fill = "black", colour = "black",
position = "identity", alpha = 0.3)
scale_y_continuous(name = "Count", breaks = seq(0,290, 10),
sec.axis = sec_axis(~.x/800, name = "TS density"))
CodePudding user response:
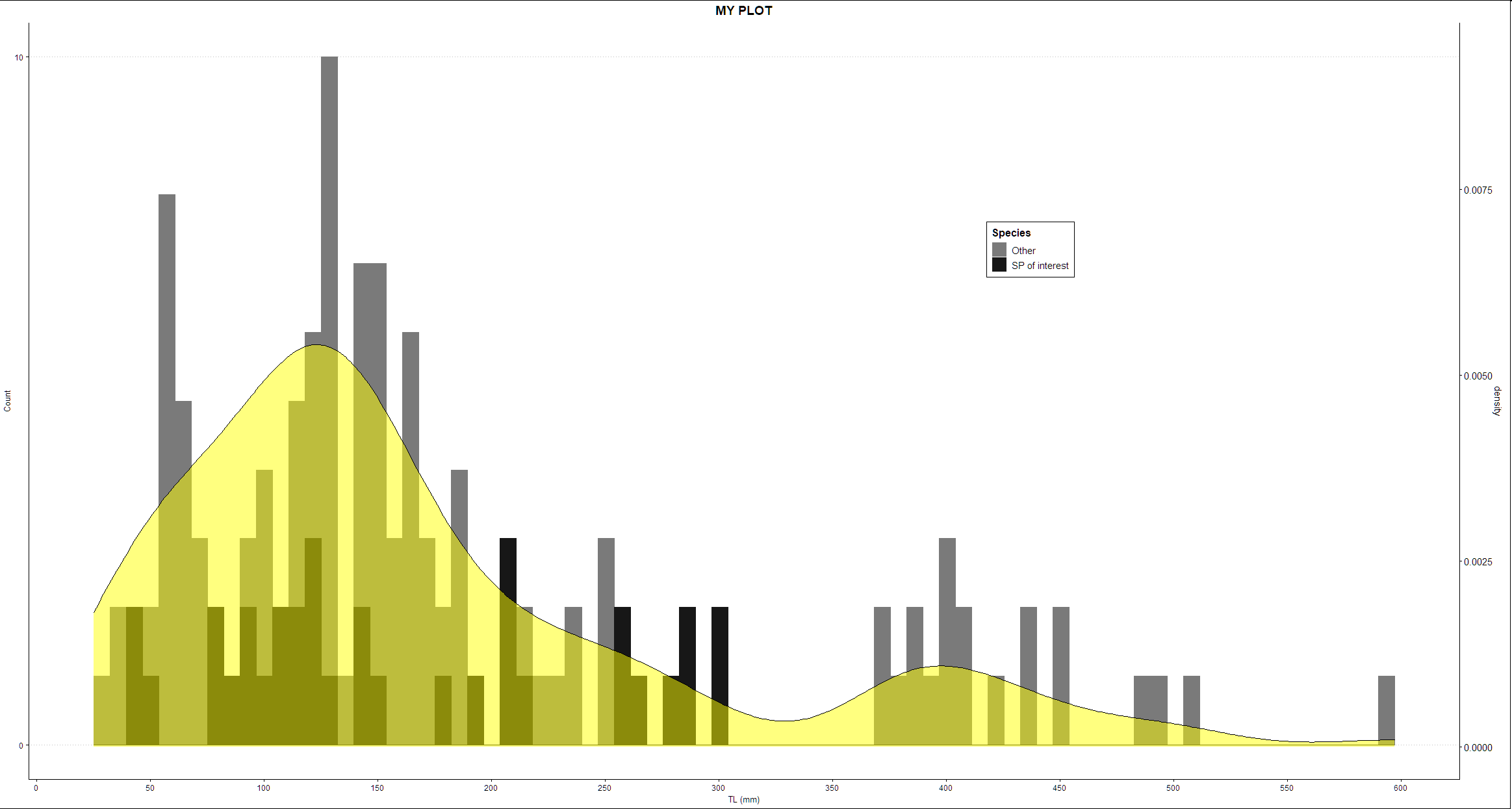
Here is an alternative using ggpubr and cowplot:
library(ggthemes)
library(ggplot2)
p_hist <- ggplot(df, aes(x=mm, color = YP, fill = YP))
geom_histogram(bins = 80, position="identity")
scale_x_continuous(name = "TL (mm)", breaks = seq(0, 800, 50))
scale_y_continuous(name = "Count", breaks = seq(0,290, 10))
scale_fill_manual(name = "Species",
labels = c("Other", "SP of interest"),
values = c("gray48", "gray9"))
scale_color_manual(name = "Species",
labels = c("Other", "SP of interest"),
values = c("gray48", "gray9"))
ggtitle("MY PLOT")
theme_clean()
theme(plot.title = element_text(hjust = 0.5), legend.position = c(0.7, 0.7))
# 2. Create the density plot with y-axis on the right
# Remove x axis elements
library(ggpubr)
library(cowplot)
p_density <- ggdensity(
df, x = "mm",
fill = "yellow")
scale_y_continuous(expand = expansion(mult = c(0.08, 0.8)), position = "right")
theme_half_open(11, rel_small = 1)
rremove("x.axis")
rremove("xlab")
rremove("x.text")
rremove("x.ticks")
rremove("legend")
aligned_plots <- align_plots(p_hist, p_density, align="hv", axis="tblr")
ggdraw(aligned_plots[[1]]) draw_plot(aligned_plots[[2]])