Reproducible Dataset
This is my data:
#### Data for Stack ####
stack <- structure(list(Mins_Work = c(435L, 350L, 145L, 135L, 15L, 60L,
60L, 390L, 395L, 395L, 315L, 80L, 580L, 175L, 545L, 230L, 435L,
370L, 255L, 515L, 330L, 65L, 115L, 550L, 420L, 45L, 266L, 196L,
198L, 220L, 17L, 382L, 0L, 180L, 343L, 207L, 263L, 332L, 0L,
0L, 259L, 417L, 282L, 685L, 517L, 111L, 64L, 466L, 499L, 460L,
269L, 300L, 427L, 301L, 436L, 342L, 229L, 379L, 102L, 146L, NA,
94L, 345L, 73L, 204L, 512L, 113L, 135L, 458L, 493L, 552L, 108L,
335L, 395L, 508L, 546L, 396L, 159L, 325L, 747L, 650L, 377L, 461L,
669L, 186L, 220L, 410L, 708L, 409L, 515L, 413L, 166L, 451L, 660L,
177L, 192L, 191L, 461L, 637L, 297L, 601L, 586L, 270L, 479L, 0L,
480L, 397L, 174L, 111L, 0L, 610L, 332L, 345L, 423L, 160L, 611L,
0L, 345L, 550L, 324L, 427L, 505L, 632L, 560L, 230L, 495L, 235L,
522L, 654L, 465L, 377L, 260L, 572L, 612L, 594L, 624L, 237L, 0L,
38L, 409L, 634L, 292L, 706L, 399L, 568L, 0L, 694L, 298L, 616L,
553L, 581L, 423L, 636L, 623L, 338L, 345L, 521L, 438L, 504L, 600L,
616L, 656L, 285L, 474L, 688L, 278L, 383L, 535L, 363L, 470L, 457L,
303L, 123L, 363L, 329L, 513L, 636L, 421L, 220L, 430L, 428L, 536L,
156L, 615L, 429L, 103L, 332L, 250L, 281L, 248L, 435L, 589L, 515L,
158L, 0L, 649L, 427L, 193L, 225L, 0L, 280L, 163L, 536L, 301L,
406L, 230L, 519L, 0L, 303L, 472L, 392L, 326L, 368L, 405L, 515L,
308L, 259L, 769L, 93L, 517L, 261L, 420L, 248L, 265L, 834L, 313L,
131L, 298L, 134L, 385L, 648L, 529L, 487L, 533L, 641L, 429L, 339L,
508L, 560L, 439L, 381L, 397L, 692L, NA), Coffee_Cups = c(3L,
0L, 2L, 6L, 4L, 5L, 3L, 3L, 2L, 2L, 3L, 1L, 1L, 3L, 2L, 2L, 0L,
1L, 1L, 4L, 4L, 3L, 0L, 1L, 3L, 0L, 0L, 0L, 0L, 2L, 0L, 1L, 2L,
3L, 2L, 2L, 4L, 3L, 6L, 6L, 3L, 4L, 6L, 8L, 3L, 5L, 0L, 2L, 2L,
8L, 6L, 4L, 6L, 4L, 4L, 2L, 6L, 6L, 5L, 1L, 3L, 1L, 5L, 4L, 6L,
5L, 0L, 6L, 6L, 4L, 4L, 2L, 2L, 6L, 6L, 7L, 3L, 3L, 0L, 5L, 7L,
6L, 3L, 5L, 3L, 3L, 1L, 9L, 9L, 3L, 3L, 6L, 6L, 6L, 3L, 0L, 7L,
6L, 6L, 3L, 9L, 3L, 8L, 8L, 3L, 3L, 7L, 6L, 3L, 3L, 3L, 6L, 6L,
6L, 1L, 9L, 3L, 3L, 2L, 6L, 3L, 6L, 9L, 6L, 8L, 9L, 6L, 6L, 6L,
0L, 3L, 0L, 3L, 3L, 6L, 3L, 0L, 9L, 3L, 0L, 2L, 0L, 6L, 6L, 6L,
3L, 6L, 3L, 9L, 3L, 0L, 0L, 6L, 3L, 3L, 3L, 3L, 6L, 0L, 6L, 3L,
3L, 5L, 5L, 3L, 0L, 6L, 4L, 2L, 0L, 2L, 4L, 0L, 6L, 4L, 4L, 2L,
2L, 0L, 9L, 6L, 3L, 6L, 6L, 9L, 0L, 6L, 6L, 6L, 6L, 6L, 6L, 3L,
3L, 0L, 9L, 6L, 3L, 6L, 3L, 6L, 1L, 6L, 6L, 6L, 6L, 6L, 1L, 3L,
9L, 6L, 3L, 6L, 9L, 3L, 5L, 6L, 3L, 0L, 6L, 3L, 3L, 5L, 0L, 6L,
3L, 5L, 3L, 0L, 6L, 7L, 3L, 6L, 6L, 6L, 6L, 3L, 5L, 6L, 7L, 6L,
6L, 4L, 3L)), class = "data.frame", row.names = c(NA, -244L))
Solution So Far
I'm trying to cut my coffee data into three groups, a low group, a medium group, and a high group. Here is how I tried doing so:
#### Load Libraries ####
library(tidyverse)
library(ggpubr)
#### Transform Data: Coffee ####
coffee_labels <- c("Low", "Medium", "High") # labels
range(stack$Coffee_Cups) # get range for split
coffee_breaks <- seq(from = 0,
to = 9,
by = 3) # split from 0 to 9 in 3 pt intervals
coffee_transform <- cut(x= stack$Coffee_Cups,
labels = coffee_labels,
breaks = coffee_breaks) # add labels and breaks
stack_transform <- stack %>%
mutate(coffee_level = coffee_transform) # mutate to add to data
tail(stack_transform$coffee_level, 30) # check transform
Problem
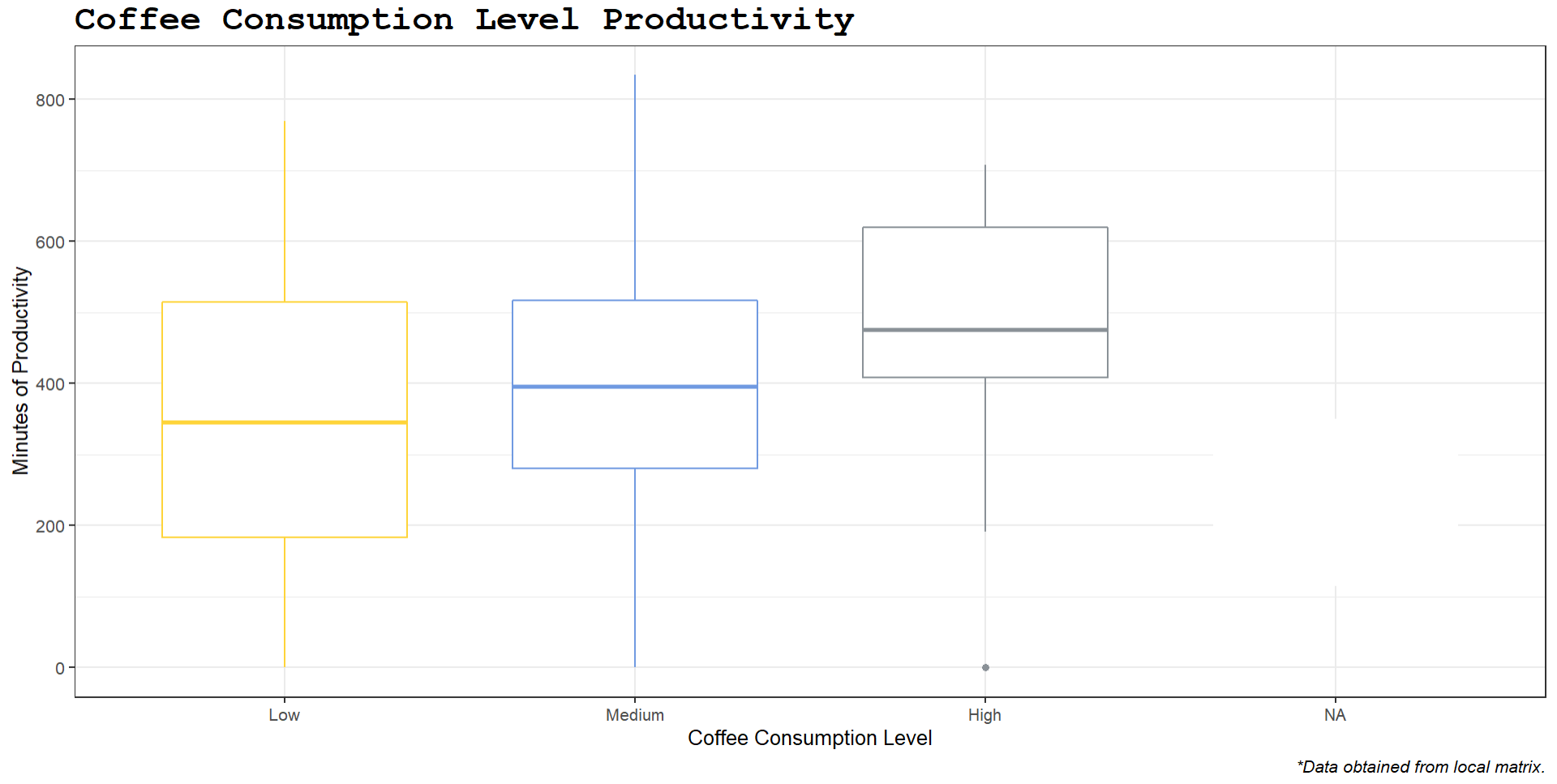
However, when I print the tail command at the end, I get these NA values, which I assume is from an improper cut:
[1] Low Medium Medium Low <NA> Medium Low Low Medium <NA> Medium
[12] Low Medium Low <NA> Medium High Low Medium Medium Medium Medium
[23] Low Medium Medium High Medium Medium Medium Low
Levels: Low Medium High
I looked and those values correspond to my coffee consumption equaling zero, yet I already set the cut from 0 to 9. Naturally, when I try to make a boxplot with this, the NA levels get included, which I don't want:
#### Transform Coffee Boxplot ####
ggboxplot(stack_transform,
x="coffee_level",
y="Mins_Work",
palette = "simpsons",
color = "coffee_level",
title = "Coffee Consumption Level Productivity",
caption = "*Data obtained from local matrix.",
xlab = "Coffee Consumption Level",
ylab = "Minutes of Productivity")
theme_bw()
theme(legend.position = "none",
plot.caption = element_text(face = "italic"),
plot.title = element_text(face = "bold",
size = 18,
family = "mono"))
Question
How do I fix these NA values? I want my zero values to be included into the "low" group if possible.
CodePudding user response:
Would this work for you? It seems you are using base R, so a nested ifelse statement may be simpler:
stack$coffee_cat <- ifelse(stack$Coffee_Cups %in% 0:3, "Low",
ifelse(stack$Coffee_Cups %in% 4:6, "Medium",
ifelse(stack$Coffee_Cups %in% 7:9, "High", NA)))
Output
# Mins_Work Coffee_Cups coffee_cat
#1 435 3 Low
#2 350 0 Low
#3 145 2 Low
#4 135 6 Medium
#5 15 4 Medium
#6 60 5 Medium
case_when would be a dplyr alternative:
stack %>% mutate(coffee_level = case_when(Coffee_Cups %in% 0:3 ~ "Low",
Coffee_Cups %in% 4:6 ~ "Medium",
Coffee_Cups %in% 7:9 ~ "High"))
To include the zeros in cutting, you could also use the Hmisc::cut2function:
stack$coffee_Hmisc <- factor(Hmisc::cut2(stack$Coffee_Cups, g = 3), labels = coffee_labels)