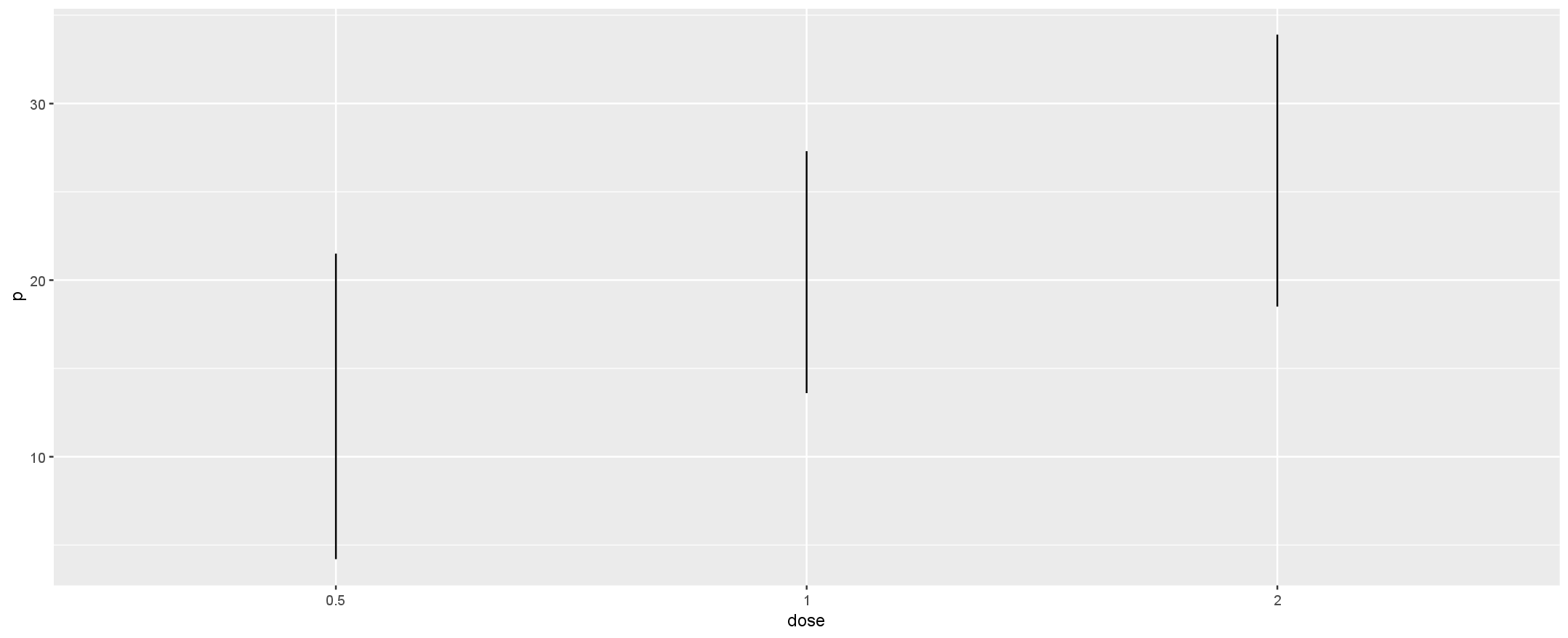
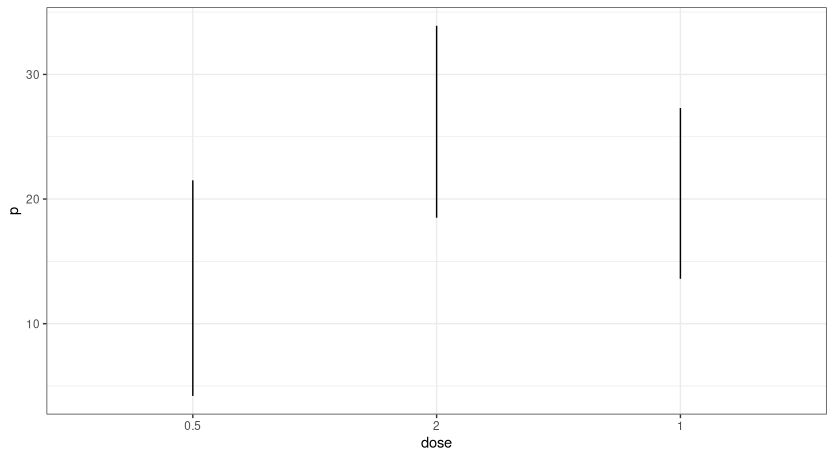
Is it possible to order line plots in R by the distance between their lower and upper value and have them ordered from greatest distance to least?
Code:
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
library(dplyr)
df.summary <- df %>%
group_by(dose) %>%
summarise(
lower = min(len), upper = max(len), p = mean(len),
)
f <- ggplot(
df.summary,
aes(x = dose, y = p, ymin = lower, ymax = upper)
)
f geom_linerange()
The goal is to have them ranked from greatest distance to least distance (left to right).
Any help at all would be greatly appreciated!
CodePudding user response:
You can use reorder()
ggplot(
df.summary,
aes(x = reorder(dose, -(upper-lower)), y = p, ymin = lower, ymax = upper))
geom_linerange()
labs(x="Dose")
CodePudding user response:
As you can see, there are a number of ways to go about this. In the below case I calculate the difference and order the data.frame according to the result. The last line sets factor levels. This is my preferred way because I keep my ggplots for plotting only and avoid doing any data manipulation if possible.
xy.summary$diff <- with(xy.summary, upper - lower)
xy.summary <- xy.summary[order(xy.summary$diff, decreasing = TRUE), ]
levels(xy.summary$dose) <- xy.summary$dose
ggplot(xy.summary, mapping = aes(x = dose, y = p, ymin = lower, ymax = upper))
theme_bw()
geom_linerange()
CodePudding user response:
this should work
df <- ToothGrowth
df$dose <- as.factor(df$dose)
head(df, 3)
library(dplyr)
df.summary <- df %>%
group_by(dose) %>%
summarise(
lower = min(len), upper = max(len), p = mean(len),
)
f <- ggplot(
df.summary,
aes(x = reorder(dose,upper-lower), y = p, ymin = lower, ymax = upper)
)
f geom_linerange()