I was tidying some data with dirty dates(different styling), I tried with * lubridate * ,however I can only clean one list at a time.
So is there anyways to convert all the date in several lists? All the lists that need to be transferred end with “T”, and, in real data I have more than 3lists, probably 30 .
The cleared data need to be “yyyy-mm”, no need of days.
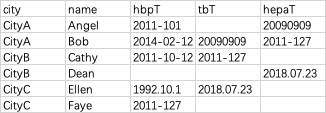
> dirtydatatest
# A tibble: 6 × 5
city name hbpT tbT hepaT
<chr> <chr> <chr> <chr> <chr>
1 CityA Angel 2011-101 NA 20090909
2 CityA Bob 2014-02-12 20090909 2011-127
3 CityB Cathy 2011-10-12 2011-127 NA
4 CityB Dean NA NA 2018.07.23
5 CityC Ellen 1992.10.1 2018.07.23 NA
6 CityC Faye 2011-127 NA NA
I used
library(tidyverse)
library(lubridate)
dirtydatatest = mutate(dirtydatatest,
tbT= ymd(tbT) ,.keep = c("all"),)
and then using stringer to change the “-”. But can only done in one list at a time.
And I tried to use
dirtydata2 %>%
select(dirtydata2, ends_with("t")) %>%
mutate_all(ymd() ,.keep = c("all"),)
But I don’t think I’ve got it right ,as in ymd(), there should be something in it...
Thank you for your time and patience.
CodePudding user response:
We can use across with ymd
library(dplyr)
library(lubridate)
dirtydataset %>%
mutate(across(ends_with("T"), ~ymd(.x, truncated = 2)))
-output
city name hbpT tbT hepaT
1 CityA Angel 2011-10-01 <NA> 2009-09-09
2 CityA Bob 2014-02-12 2009-09-09 2011-12-07
3 CityB Cathy 2011-10-12 2011-12-07 <NA>
4 CityB Dean <NA> <NA> 2018-07-23
5 CityC Ellen 1992-10-01 2018-07-23 <NA>
6 CityC Faye 2011-12-07 <NA> <NA>
With mutate_at, the syntax would be
dirtydataset %>%
mutate_at(vars(ends_with("T")), ~ ymd(.x, truncated = 2))
city name hbpT tbT hepaT
1 CityA Angel 2011-10-01 <NA> 2009-09-09
2 CityA Bob 2014-02-12 2009-09-09 2011-12-07
3 CityB Cathy 2011-10-12 2011-12-07 <NA>
4 CityB Dean <NA> <NA> 2018-07-23
5 CityC Ellen 1992-10-01 2018-07-23 <NA>
6 CityC Faye 2011-12-07 <NA> <NA>
data
dirtydataset <- structure(list(city = c("CityA", "CityA", "CityB",
"CityB", "CityC",
"CityC"), name = c("Angel", "Bob", "Cathy", "Dean", "Ellen",
"Faye"), hbpT = c("2011-101", "2014-02-12", "2011-10-12", NA,
"1992.10.1", "2011-127"), tbT = c(NA, "20090909", "2011-127",
NA, "2018.07.23", NA), hepaT = c("20090909", "2011-127", NA,
"2018.07.23", NA, NA)), class = "data.frame", row.names = c("1",
"2", "3", "4", "5", "6"))
CodePudding user response:
As an alternative we could use parse_date function from parsedate package. Data from @r2evans (Many thanks).
parse_date can parse a date when you don’t know which format it is in. First it tries all ISO 8601 formats. Then it tries git’s versatile date parser. Lastly, it tries as.POSIXct.
library(dplyr)
library(parsedate)
dirtydataset %>%
mutate(across(ends_with("T"), ~parsedate::parse_date(.)))
output:
city name hbpT tbT hepaT
1 CityA Angel 2011-04-11 <NA> 2009-09-09
2 CityA Bob 2014-02-12 2009-09-09 2011-05-07
3 CityB Cathy 2011-10-12 2011-05-07 <NA>
4 CityB Dean <NA> <NA> 2018-07-23
5 CityC Ellen 1992-10-01 2018-07-23 <NA>
6 CityC Faye 2011-05-07 <NA> <NA>
CodePudding user response:
First, I think across works well here to convert multiple columns to Date-class (and mutate_all and mutate_at have been superseded by it):
dirtydataset %>%
mutate(across(ends_with("T"), ymd))
# # A tibble: 6 x 5
# city name hbpT tbT hepaT
# <chr> <chr> <date> <date> <date>
# 1 CityA Angel 2011-10-01 NA 2009-09-09
# 2 CityA Bob 2014-02-12 2009-09-09 2011-12-07
# 3 CityB Cathy 2011-10-12 2011-12-07 NA
# 4 CityB Dean NA NA 2018-07-23
# 5 CityC Ellen 1992-10-01 2018-07-23 NA
# 6 CityC Faye 2011-12-07 NA NA
As to your mention of "need to be “yyyy-mm”, no need of days", then you can convert back to strings if you'd like, using format, though you lose some capabilities on it:
dirtydataset %>%
mutate(across(ends_with("T"), ~ format(ymd(.), format = "%Y-%m")))
# # A tibble: 6 x 5
# city name hbpT tbT hepaT
# <chr> <chr> <chr> <chr> <chr>
# 1 CityA Angel 2011-10 NA 2009-09
# 2 CityA Bob 2014-02 2009-09 2011-12
# 3 CityB Cathy 2011-10 2011-12 NA
# 4 CityB Dean NA NA 2018-07
# 5 CityC Ellen 1992-10 2018-07 NA
# 6 CityC Faye 2011-12 NA NA
Data
dirtydataset <- structure(list(city = c("CityA", "CityA", "CityB", "CityB", "CityC", "CityC"), name = c("Angel", "Bob", "Cathy", "Dean", "Ellen", "Faye"), hbpT = c("2011-101", "2014-02-12", "2011-10-12", NA, "1992.10.1", "2011-127"), tbT = c(NA, "20090909", "2011-127", NA, "2018.07.23", NA), hepaT = c("20090909", "2011-127", NA, "2018.07.23", NA, NA)), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA, -6L))
