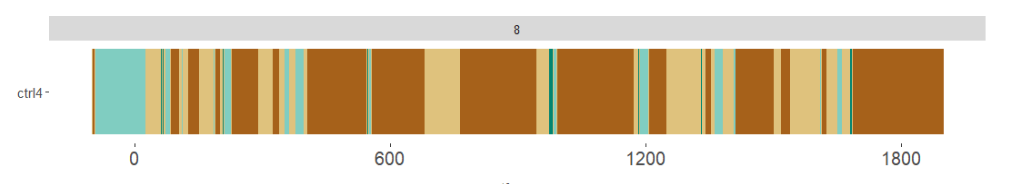
I have a question about axis label positioning that I haven't been able to resolve.
I am using geom_tile plotting for a time series data. I am basically plotting the zone that the animal was in relative to the time course of the experiment. Below is an example df:
time speed zone
<dbl> <dbl> <chr>
1 0 NA eating
2 0.3 NA eating
3 0.334 NA eating
4 0.368 NA nest
5 0.401 NA nest
6 0.434 NA nest
The plotting works just fine, however, when I get the graph the x axis begins and finishes outside of the actual experiment time. All experiments start precisely at 0 and finish precisely at 1800. However, the graph looks like this:
Can anyone help me with that? If I set the xlim (0, 1800) it works fine, but then the breaks are at 500, 1000 and 1500, whereas I would like them at 600, 1200 and 1800.
Here is the code I'm using to plot:
ggplot(df, aes(x = time, y = animal, fill=zone, width = 200))
geom_tile()
facet_wrap(.~week, ncol=1)
scale_fill_brewer(palette = "BrBG")
scale_x_time(expand = c(0, 0))
scale_y_discrete(expand = c(0, 0))
Thank you in advance!
CodePudding user response:
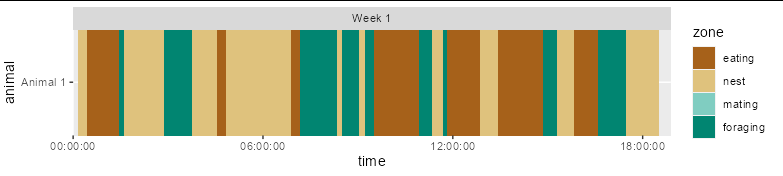
geom_tile isn't the right tool here. It will give you a fixed width centred on the time, but what you need is a variable width rectangle with a left edge at time and a right edge at the next time, so you should perhaps use geom_rect:
ggplot(df, aes(x = time, y = animal, fill = zone))
geom_rect(aes(xmin = time, xmax = c(time[-1], tail(time, 1)),
ymin = as.numeric(factor(animal)) - 0.5,
ymax = as.numeric(factor(animal)) 0.5))
facet_wrap(.~week, ncol=1)
scale_fill_brewer(palette = "BrBG", drop = FALSE)
scale_x_time(expand = c(0, 0), breaks = 60 * c(0, 360, 720, 1080),
limits = c(0, 68000))
scale_y_discrete(expand = c(0, 0))
Note you didn't have enough sample data to demonstrate this properly, so I created some:
set.seed(1)
df <- data.frame(time = cumsum(runif(50, 0, 2500)),
zone = factor(sample(c('eating', 'nest', 'foraging'), 50, TRUE),
c('eating', 'nest', 'mating', 'foraging')),
week = "Week 1", animal = "Animal 1")
CodePudding user response:
geom_tile creates a rectangle in which x and y represent the center of the tile. This results into an x axis showing data before 0 and after 1800. Use geom_rect instead:
library(tidyverse)
df <- tibble(
time = c(0, 200,1500,1800),
zone = c("eating", "eating", "nest", "eating"),
week = 1,
animal = "ctrl4"
)
ggplot(df)
geom_rect(aes(xmin = time, xmax = lead(time), ymin = 0, ymax= 1, fill = zone))
facet_wrap(.~week, ncol=1)
scale_y_discrete(expand = c(0, 0))
scale_x_continuous(expand = c(0,0))
#> Warning: Removed 1 rows containing missing values (geom_rect).
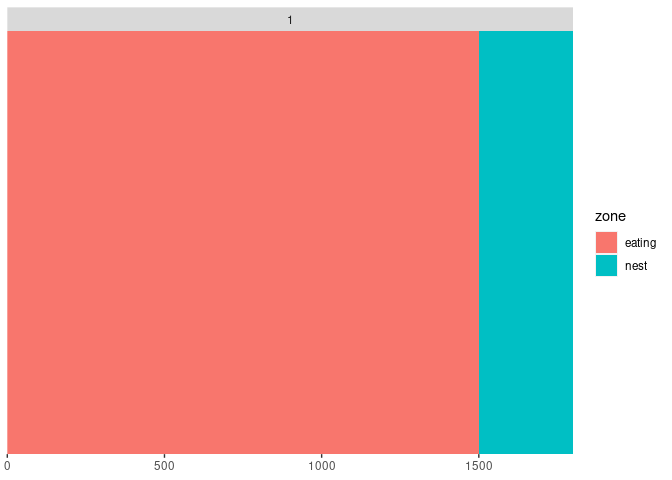
Created on 2022-05-20 by the reprex package (v2.0.0)