I have a big dataframe that looks like this:
> dput(sb_data_omit_1950[sample(nrow(sb_data_omit_1950), 50),])
structure(list(lat = c("56", "61", "57", "59", "58", "56", "58",
"65", "59", "65", "63", "65", "56", "59", "59", "57", "59", "60",
"56", "57", "60", "65", "64", "63", "63", "59", "59", "65", "59",
"58", "63", "59", "64", "59", "58", "59", "63", "56", "58", "59",
"57", "55", "58", "64", "62", "60", "57", "58", "60", "66"),
long = c(18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18,
18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18,
18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18, 18,
18, 18, 18, 18, 18, 18, 18, 18), date = c("2009-02-07", "1995-03-04",
"2007-01-28", "2010-03-28", "2010-04-01", "2018-02-22", "2017-03-24",
"2014-04-16", "1983-03-20", "2016-04-02", "2020-04-14", "2020-04-02",
"2005-03-25", "2003-03-22", "2016-04-02", "2006-03-19", "2009-04-05",
"2009-01-22", "2016-03-05", "2013-02-23", "2017-03-17", "2020-03-25",
"2021-03-27", "2008-04-08", "2018-04-10", "1984-04-04", "2005-01-29",
"2019-04-03", "1983-04-10", "2006-03-26", "2010-03-29", "2006-03-18",
"2014-05-06", "2010-01-23", "2006-03-26", "2014-02-25", "2008-04-16",
"2021-02-16", "2011-03-30", "2013-03-07", "1975-03-22", "2015-02-01",
"2013-03-21", "2011-04-07", "2021-04-06", "2021-02-02", "2000-03-19",
"1983-02-26", "2010-04-03", "2017-03-28"), julian_day = c(38,
63, 28, 87, 91, 53, 83, 106, 79, 93, 105, 93, 84, 81, 93,
78, 95, 22, 65, 54, 76, 85, 86, 99, 100, 95, 29, 93, 100,
85, 88, 77, 126, 23, 85, 56, 107, 47, 89, 66, 81, 32, 80,
97, 96, 33, 79, 57, 93, 87), year = c(2009L, 1995L, 2007L,
2010L, 2010L, 2018L, 2017L, 2014L, 1983L, 2016L, 2020L, 2020L,
2005L, 2003L, 2016L, 2006L, 2009L, 2009L, 2016L, 2013L, 2017L,
2020L, 2021L, 2008L, 2018L, 1984L, 2005L, 2019L, 1983L, 2006L,
2010L, 2006L, 2014L, 2010L, 2006L, 2014L, 2008L, 2021L, 2011L,
2013L, 1975L, 2015L, 2013L, 2011L, 2021L, 2021L, 2000L, 1983L,
2010L, 2017L), decade = c("2000-2009", "1990-1999", "2000-2009",
"2010-2019", "2010-2019", "2010-2019", "2010-2019", "2010-2019",
"1980-1989", "2010-2019", "2020-2029", "2020-2029", "2000-2009",
"2000-2009", "2010-2019", "2000-2009", "2000-2009", "2000-2009",
"2010-2019", "2010-2019", "2010-2019", "2020-2029", "2020-2029",
"2000-2009", "2010-2019", "1980-1989", "2000-2009", "2010-2019",
"1980-1989", "2000-2009", "2010-2019", "2000-2009", "2010-2019",
"2010-2019", "2000-2009", "2010-2019", "2000-2009", "2020-2029",
"2010-2019", "2010-2019", "1970-1979", "2010-2019", "2010-2019",
"2010-2019", "2020-2029", "2020-2029", "2000-2009", "1980-1989",
"2010-2019", "2010-2019"), time = c(15L, 14L, 15L, 16L, 16L,
16L, 16L, 16L, 13L, 16L, 17L, 17L, 15L, 15L, 16L, 15L, 15L,
15L, 16L, 16L, 16L, 17L, 17L, 15L, 16L, 13L, 15L, 16L, 13L,
15L, 16L, 15L, 16L, 16L, 15L, 16L, 15L, 17L, 16L, 16L, 12L,
16L, 16L, 16L, 17L, 17L, 15L, 13L, 16L, 16L), lat_grouped = c("1",
"2", "1", "1", "1", "1", "1", "3", "1", "3", "2", "3", "1",
"1", "1", "1", "1", "2", "1", "1", "2", "3", "2", "2", "2",
"1", "1", "3", "1", "1", "2", "1", "2", "1", "1", "1", "2",
"1", "1", "1", "1", "1", "1", "2", "2", "2", "1", "1", "2",
"3")), row.names = c(21286L, 5843L, 16479L, 24246L, 24483L,
40513L, 39121L, 33554L, 2704L, 37376L, 48602L, 48008L, 12473L,
9593L, 37380L, 14123L, 22712L, 21155L, 36663L, 29846L, 38722L,
47518L, 51286L, 20119L, 42528L, 3132L, 11764L, 44966L, 2874L,
14406L, 24290L, 14081L, 33634L, 23125L, 14393L, 31981L, 20790L,
50057L, 26126L, 30068L, 1381L, 34000L, 30253L, 26612L, 51918L,
49677L, 7640L, 2677L, 24745L, 39308L), class = "data.frame")
> head(df)
lat long date julian_day year decade time lat_grouped
24 59 18 1951-03-22 81 1951 1950-1959 10 1
25 59 18 1951-04-08 98 1951 1950-1959 10 1
26 55 18 1952-02-03 34 1952 1950-1959 10 1
27 59 18 1952-03-08 68 1952 1950-1959 10 1
28 59 18 1953-02-22 53 1953 1950-1959 10 1
29 63 18 1953-03-12 71 1953 1950-1959 10 2
From this data, I would like to count the amount of observations per julian day (variable julian_day) in a given decade (two variables decade or time which is a numerical translation of the first one) and plot it against the data of other decades on the same graphic.
Until now, I have managed to plot the count of observations for all decades together using this code:
df %>%
ggplot(aes(x=julian_day))
geom_histogram(color="darkblue", fill="white", bins=152)
xlim(0, 155)
xlab("Day n°") ylab("Count")
I have the intuition that I should use a group_by(time) but cannot manage to plot some selected groups.
The output should look like multiple gaussian curves plotted on the same diagram.
Can anyone help? Thank you very much, if any information is missing I can edit my post :)
CodePudding user response:
I am not 100% sure what your final output should be, but if you want all the histograms on one plot:
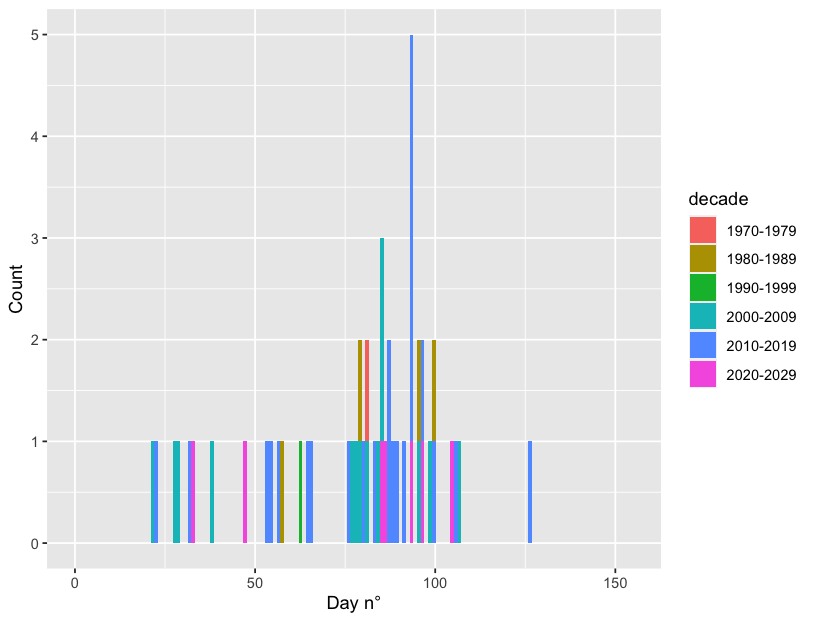
df %>%
ggplot(aes(x=julian_day, fill = decade))
geom_histogram(bins=152)
xlim(0, 155)
xlab("Day n°") ylab("Count")
If you separate histograms on the same figure:
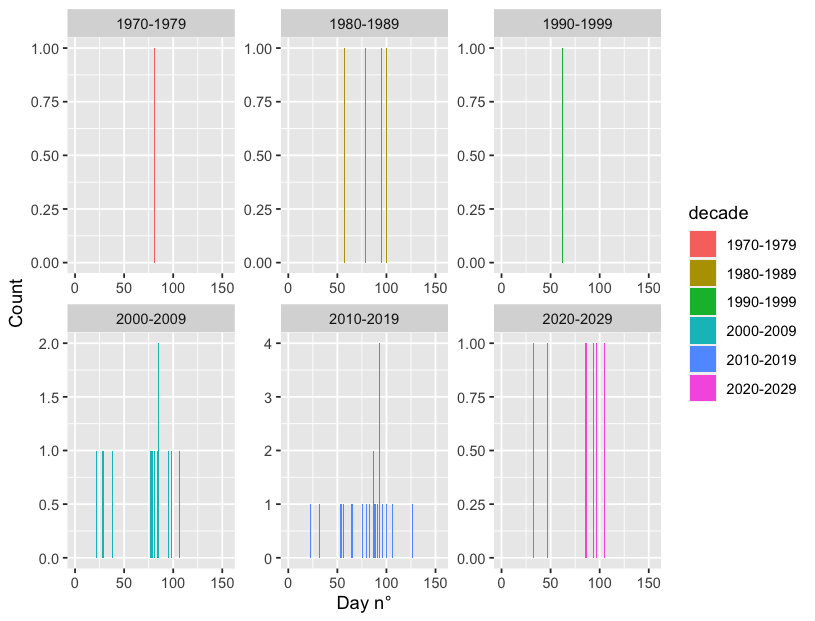
df %>%
ggplot(aes(x=julian_day, fill = decade))
geom_histogram(bins=152)
xlim(0, 155)
xlab("Day n°") ylab("Count")
facet_wrap(~decade, scales = "free")
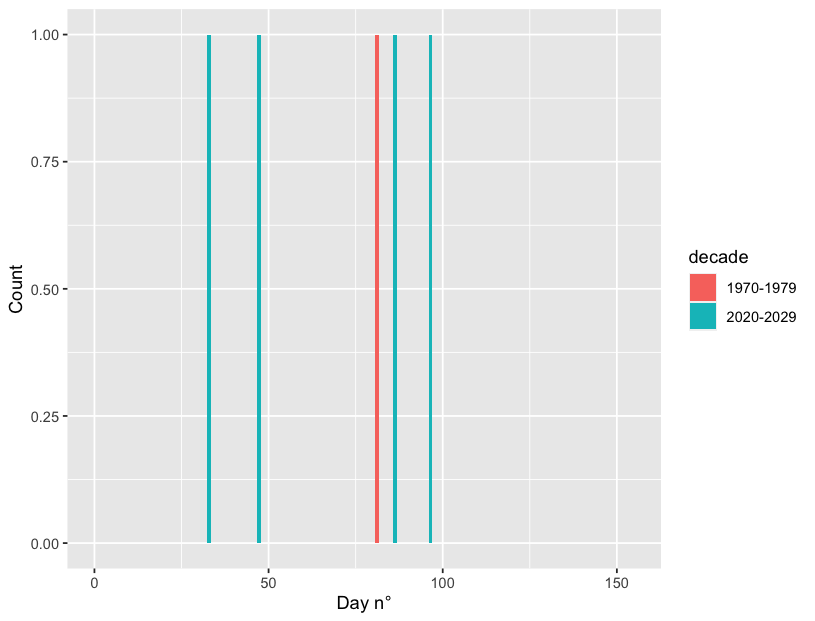
If you only want to select certain decades, you can add a filter() argument. The easiest way in this situation would be to filter by year, since it is numeric:
# first and last decade
keeps <- c(min(df$year), max(df$year))
# or any decade by referencing a year within that decade
# keeps <- c(2009, 1985)
df %>% filter(year %in% keeps) %>%
ggplot(aes(x=julian_day, fill = decade))
geom_histogram(bins=152)
xlim(0, 155)
xlab("Day n°") ylab("Count")
CodePudding user response:
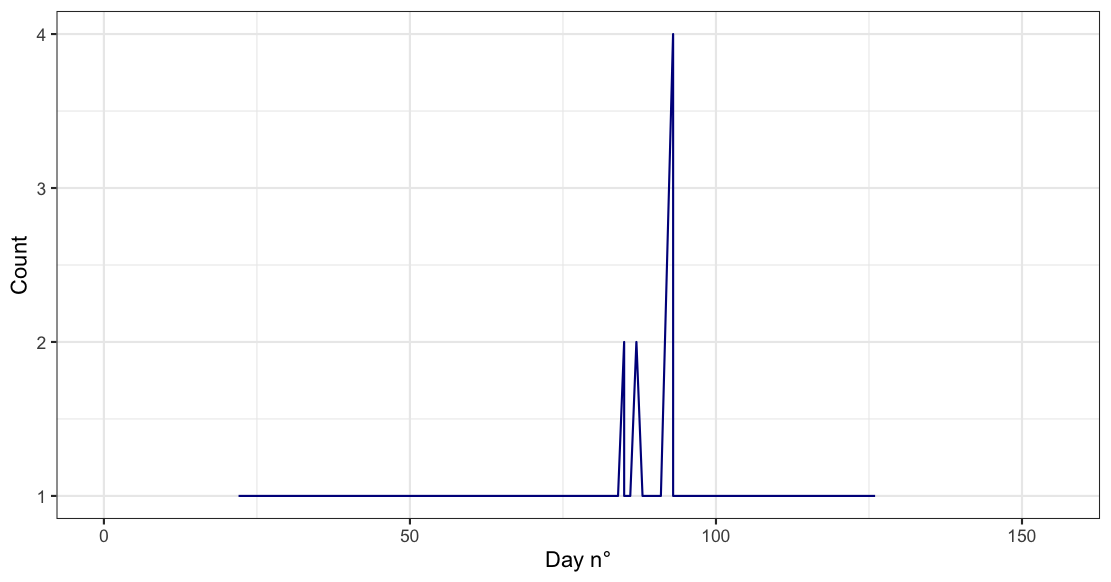
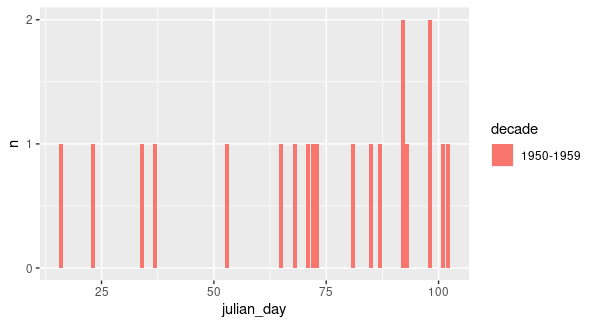
Maybe you want something like this:
df %>%
count(decade, julian_day) %>%
ggplot(aes(x=julian_day, n))
geom_line(color="darkblue")
xlim(0, 155)
xlab("Day n°")
ylab("Count")
theme_bw()
Output:
CodePudding user response:
Update: See comments:
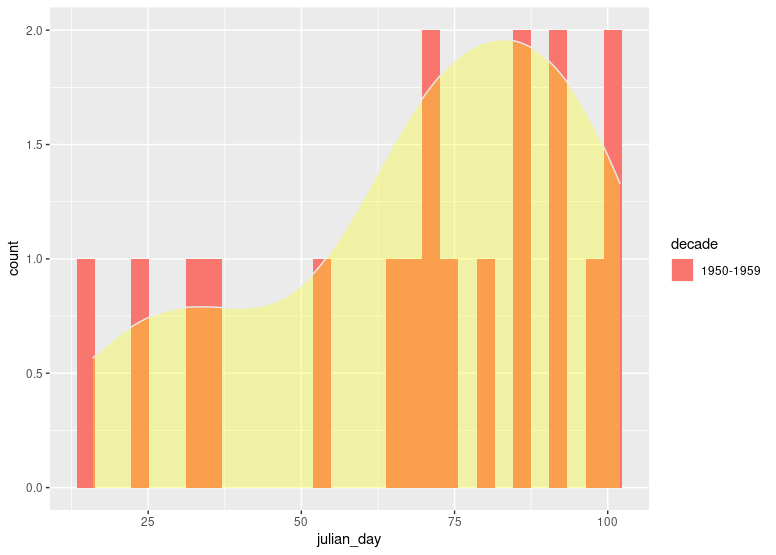
We could combine geom_histogram applying count statistics with geom_density applying density statistics:
df %>%
count(decade, julian_day) %>%
ggplot(aes(x = julian_day, fill=decade))
stat_bin(bins = 30, aes(y = ..count..))
geom_density(aes(y = ..density..*(nrow(df1)*0.8)), fill="yellow", color="#e9ecef", alpha=0.3)
library(tidyverse)
df %>%
count(decade, julian_day) %>%
ggplot(aes(x = julian_day, y=n, fill=decade))
geom_col(position= position_dodge())
scale_y_continuous(breaks = function(x) unique(floor(pretty(seq(0, (max(x) 1) * 1.1)))))