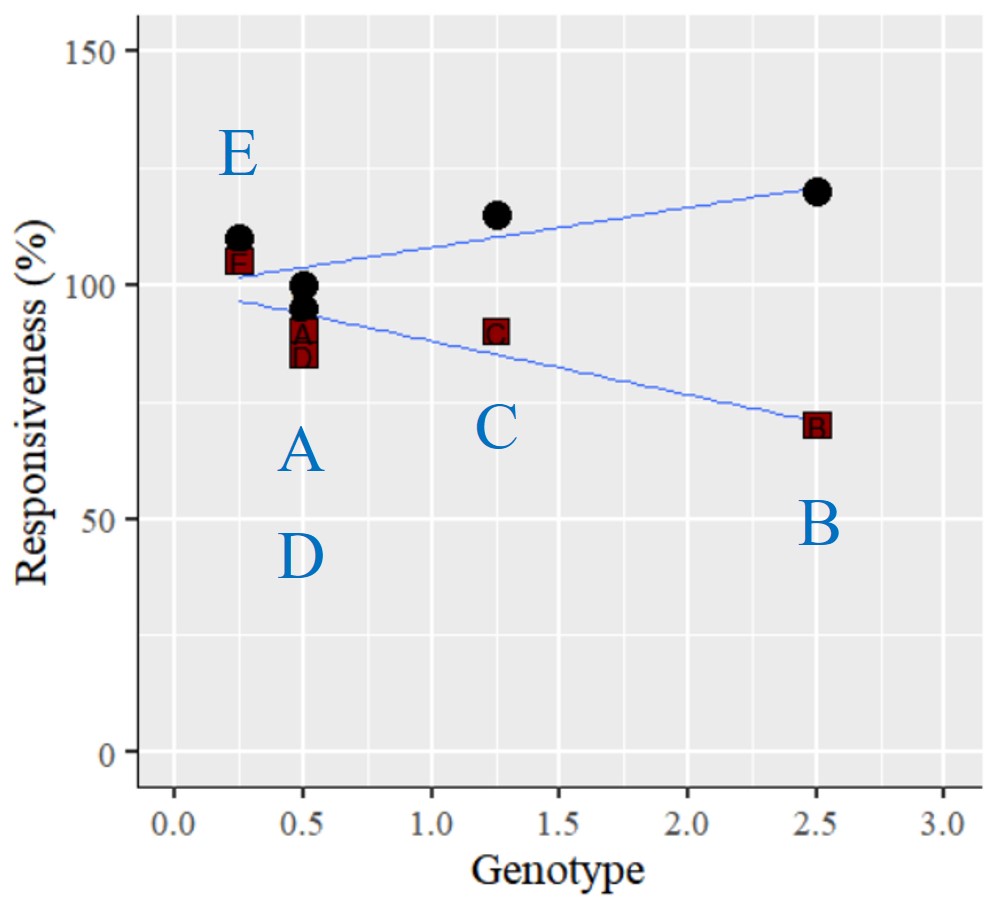
I'd like to know how to adjust the position of data label inside graph in R. This is my data and graph.
Genotype<- rep(c("A","B","C","D","E"), times=2)
Factor<- rep(c("Control","Treatment"), each=5)
Control<- c(100,120,115,95,110,90,70,90,85,105)
Reaction_norm <- c(0.5,2.5,1.3,0.5,0.3,0.5,2.5,1.3,0.5,0.3)
DataA<- data.frame(Genotype,Factor,Control,Reaction_norm)
DataA
ggplot(data=dataA, aes(x=norm, y=Control))
geom_smooth(aes(group=Factor), method=lm, level=0.95, se=FALSE, linetype=1,
size=0.5, formula=y~x)
geom_point (aes(shape=Factor, fill=Factor), col="Black", size=5)
geom_text(aes(label=Genotype, size=NULL))
scale_fill_manual(values = c("Black","Dark red"))
scale_shape_manual(values = c(21,22))
scale_x_continuous(breaks = seq(0,3,0.5),limits = c(0,3))
scale_y_continuous(breaks = seq(0,150,50), limits = c(0,150))
labs(x="Genotype", y="Responsiveness (%)")
theme_grey(base_size=17, base_family="serif")
theme(legend.position= 'none',
axis.line= element_line(size=0.5, colour="black"))
windows(width=5.5, height=5)
The data label is placed on the inside the point, but I want to place data label in different location like blue texts.
So, how to place the data label in different location?
CodePudding user response:
Pre-calculate y position for every label and then pass aes(y = Genotype_y) to geom_text.
If you're OK with having all the labels strictly below or above each point, then pass nudge_y to geom_text, see https://ggplot2.tidyverse.org/reference/geom_text.html
CodePudding user response:
One potential option is to use the ggrepel package, e.g.
library(ggplot2)
library(ggrepel)
Genotype<- rep(c("A","B","C","D","E"), times=2)
Factor<- rep(c("Control","Treatment"), each=5)
Control<- c(100,120,115,95,110,90,70,90,85,105)
Reaction_norm <- c(0.5,2.5,1.3,0.5,0.3,0.5,2.5,1.3,0.5,0.3)
DataA <- data.frame(Genotype,Factor,Control,Reaction_norm)
DataA
#> Genotype Factor Control Reaction_norm
#> 1 A Control 100 0.5
#> 2 B Control 120 2.5
#> 3 C Control 115 1.3
#> 4 D Control 95 0.5
#> 5 E Control 110 0.3
#> 6 A Treatment 90 0.5
#> 7 B Treatment 70 2.5
#> 8 C Treatment 90 1.3
#> 9 D Treatment 85 0.5
#> 10 E Treatment 105 0.3
ggplot(data=DataA, aes(x=Reaction_norm, y=Control))
geom_smooth(aes(group=Factor), method=lm, level=0.95, se=FALSE, linetype=1,
size=0.5, formula=y~x)
geom_point(aes(shape=Factor, fill=Factor), col="Black", size=5)
geom_label_repel(aes(label=Genotype),
min.segment.length = 0)
scale_fill_manual(values = c("Black","Dark red"))
scale_shape_manual(values = c(21,22))
scale_x_continuous(breaks = seq(0,3,0.5),limits = c(0,3))
scale_y_continuous(breaks = seq(0,150,50), limits = c(0,150))
labs(x="Genotype", y="Responsiveness (%)")
theme_grey(base_size=17, base_family="serif")
theme(legend.position= 'none',
axis.line= element_line(size=0.5, colour="black"))
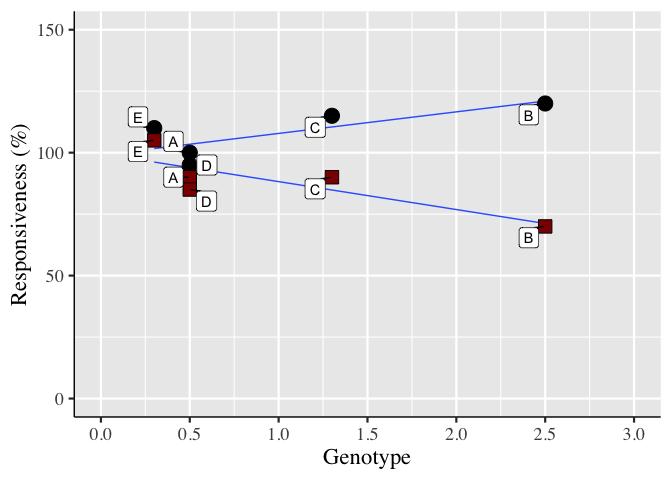
Created on 2022-05-29 by the reprex package (v2.0.1)
Or, with text instead of labels:
library(ggplot2)
library(ggrepel)
Genotype<- rep(c("A","B","C","D","E"), times=2)
Factor<- rep(c("Control","Treatment"), each=5)
Control<- c(100,120,115,95,110,90,70,90,85,105)
Reaction_norm <- c(0.5,2.5,1.3,0.5,0.3,0.5,2.5,1.3,0.5,0.3)
DataA <- data.frame(Genotype,Factor,Control,Reaction_norm)
DataA
#> Genotype Factor Control Reaction_norm
#> 1 A Control 100 0.5
#> 2 B Control 120 2.5
#> 3 C Control 115 1.3
#> 4 D Control 95 0.5
#> 5 E Control 110 0.3
#> 6 A Treatment 90 0.5
#> 7 B Treatment 70 2.5
#> 8 C Treatment 90 1.3
#> 9 D Treatment 85 0.5
#> 10 E Treatment 105 0.3
ggplot(data=DataA, aes(x=Reaction_norm, y=Control))
geom_smooth(aes(group=Factor), method=lm, level=0.95, se=FALSE, linetype=1,
size=0.5, formula=y~x)
geom_point(aes(shape=Factor, fill=Factor), col="Black", size=5)
geom_text_repel(aes(label=Genotype),
min.segment.length = 0,
box.padding = 2,
max.overlaps = 100,
size = 8)
scale_fill_manual(values = c("Black","Dark red"))
scale_shape_manual(values = c(21,22))
scale_x_continuous(breaks = seq(0,3,0.5),limits = c(0,3))
scale_y_continuous(breaks = seq(0,150,50), limits = c(0,150))
labs(x="Genotype", y="Responsiveness (%)")
theme_grey(base_size=17, base_family="serif")
theme(legend.position= 'none',
axis.line= element_line(size=0.5, colour="black"))

Created on 2022-05-29 by the reprex package (v2.0.1)