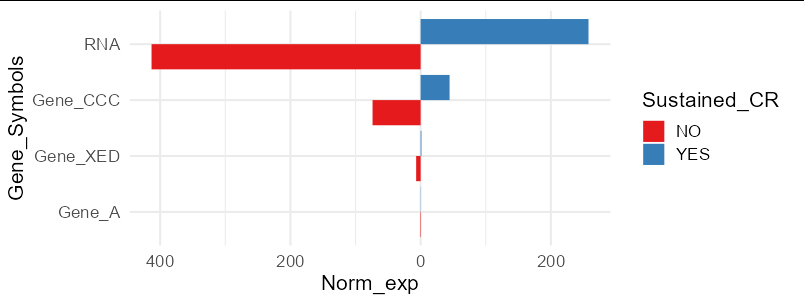
I am trying to draw a barplot on either side of the y-axis (atleast present in "YES" or "NO") instead of stacking both "YES" and "NO" together. Firstly I convert the data.frame to longer format > Calculated mean > ggplot2.
When I ran the ggplot2 code, the barplot is stacked with both "YES" and "NO" together (see screenshot below). I have provided example data that I am using to plot. Basically, I would like to view as expected plot. Is there any additional step that I am missing? Please assist.
dput(Data)
structure(list(SampleID = c("Sample_1", "Sample_2", "Sample_3",
"Sample_4", "Sample_5", "Sample_6"), Sustained_CR = c("NO", "NO",
"NO", "YES", "YES", "YES"), Gene_A = c(2.245937679, 0, 0, 1.128343065,
0, 0), Gene_CCC = c(83.09969414, 16.10799215, 122.2752332, 57.54549633,
13.12780895, 62.70159708), Gene_XED = c(6.737813038, 1.006749509,
12.80978634, 1.128343065, 0, 2.508063883), RNA = c(580.5748901,
312.0923478, 347.0287571, 292.2408539, 355.7636226, 125.4031942
)), row.names = c("Sample_1", "Sample_2", "Sample_3", "Sample_4",
"Sample_5", "Sample_6"), class = "data.frame")
library(tidyverse)
Data_long <- Data[, -c(1)] %>% pivot_longer(cols = -"Sustained_CR",names_to="Gene_Symbols",values_to="Normalized expression values")
sum(is.na(Data_long))
head(Data_long)
## Calculate the mean based on the Gene_Symbols
Data_mean <- Data_long %>% group_by(Gene_Symbols, Sustained_CR) %>% summarize(Norm_exp=mean(`Normalized expression values`, na.rm = T))
print(Data_mean)
Gene_Symbols Sustained_CR Norm_exp
<chr> <chr> <dbl>
1 Gene_A NO 0.749
2 Gene_A YES 0.376
3 Gene_CCC NO 73.8
4 Gene_CCC YES 44.5
5 Gene_XED NO 6.85
6 Gene_XED YES 1.21
7 RNA NO 413.
8 RNA YES 258.
ggplot(Data_mean, aes(reorder(Gene_Symbols, -Norm_exp), Norm_exp, fill = Sustained_CR))
geom_bar(stat = "identity")
coord_flip()
xlab("Gene_Symbols")
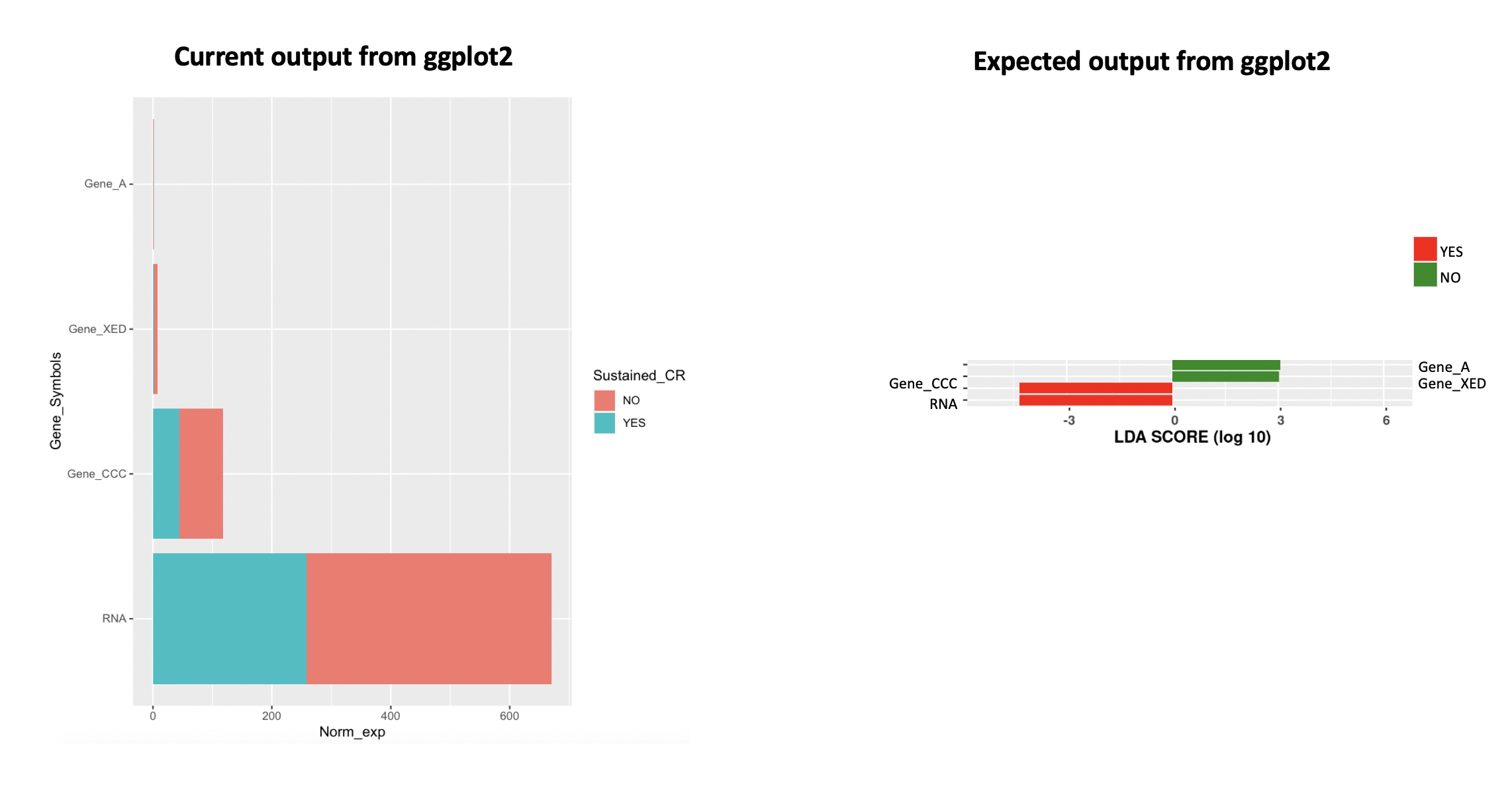
Actually, I dont want the stacked barplot on either side. The bar should be either present in "YES" or "NO" only.
Thank You,
Toufiq
CodePudding user response:
You need to make the Norm_exp values negative when Sustained_CR is "NO":
Data_mean %>%
mutate(Norm_exp = ifelse(Sustained_CR == "YES", Norm_exp, -Norm_exp)) %>%
ggplot(aes(reorder(Gene_Symbols, -Norm_exp), Norm_exp, fill = Sustained_CR))
geom_col(position = "dodge")
coord_flip()
xlab("Gene_Symbols")
scale_fill_brewer(palette = "Set1")
theme_minimal(base_size = 16)
scale_y_continuous(labels = abs)