This is the data:
tmp <- structure(list(id = c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3,
3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3), perc = c(1, 0.996059244,
1.001618656, 0.997829726, 0.915108419, 0.92539353, 0.047364961,
0.017640387, 0.036429124, 0.045454043, -0.006316982, 0.025515999,
-0.011243315, 0.00177058, 0.083566957, 0.018730071, 10.893466942,
0.635321677, 0.486662427, 0.421685776, 0.398957515, 0.397167489,
0.406132297, 0.42004932, 0.436092126, 0.451437885, 0.469267847,
0.484069249, 0.496775105, 0.510017824, 0.517280558, 1, 0.970310965,
0.920600257, 0.801496781, 0.635352677, 0.351879201, 0.133918706,
0.020005058, -0.003554937, -0.027281619, -0.030216871, -0.035568669,
-0.018927467, -0.054635806, -0.023042942, -0.049607356, 1), breakpoint = c(1,
1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 1)), class = c("grouped_df", "tbl_df", "tbl", "data.frame"
), row.names = c(NA, -48L), groups = structure(list(id = c(1,
2, 3), .rows = structure(list(1:16, 17:32, 33:48), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -3L), .drop = TRUE))
I want to count the values greater than 0.8 and record it to a new column, group by ID, so far I'm able to use ifelse to get 0 and 1 and store in another column, but get stuck after that... What I tried:
tmp <- tmp %>%
dplyr::group_by(id)%>%
dplyr::mutate(breakpoint = ifelse(perc >= 0.8, 1,0))
This is my desired out:
structure(list(ID = c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L), percentage = c(1, 0.996059244, 1.001618656,
0.997829726, 0.915108419, 0.92539353, 0.047364961, 0.017640387,
0.036429124, 0.045454043, -0.006316982, 0.025515999, -0.011243315,
0.00177058, 0.083566957, 0.018730071, 1, 0.893466942, 0.635321677,
0.486662427, 0.421685776, 0.398957515, 0.397167489, 0.406132297,
0.42004932, 0.436092126, 0.451437885, 0.469267847, 0.484069249,
0.496775105, 0.510017824, 0.517280558, 1, 0.970310965, 0.920600257,
0.801496781, 0.635352677, 0.351879201, 0.133918706, 0.020005058,
-0.003554937, -0.027281619, -0.030216871, -0.035568669, -0.018927467,
-0.054635806, -0.023042942, -0.049607356), breakpoint = c(NA,
NA, NA, NA, NA, 6L, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
2L, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, 4L, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA)), class = "data.frame", row.names = c(NA,
-48L))
Can someone also tell me how to display the output as code? Only know to use ctrl k...Thanks!
CodePudding user response:
tmp %>%
group_by(id) %>%
mutate(res = cumsum(breakpoint)*breakpoint,
res = res * NA ^(res != max(res)))
id perc breakpoint res
1 1 1.000000000 1 NA
2 1 0.996059244 1 NA
3 1 1.001618656 1 NA
4 1 0.997829726 1 NA
5 1 0.915108419 1 NA
6 1 0.925393530 1 6
7 1 0.047364961 0 NA
8 1 0.017640387 0 NA
9 1 0.036429124 0 NA
10 1 0.045454043 0 NA
11 1 -0.006316982 0 NA
12 1 0.025515999 0 NA
13 1 -0.011243315 0 NA
14 1 0.001770580 0 NA
15 1 0.083566957 0 NA
16 1 0.018730071 0 NA
17 2 10.893466942 1 NA
18 2 0.635321677 0 NA
19 2 0.486662427 0 NA
20 2 0.421685776 0 NA
21 2 0.398957515 0 NA
22 2 0.397167489 0 NA
23 2 0.406132297 0 NA
24 2 0.420049320 0 NA
25 2 0.436092126 0 NA
26 2 0.451437885 0 NA
27 2 0.469267847 0 NA
28 2 0.484069249 0 NA
29 2 0.496775105 0 NA
30 2 0.510017824 0 NA
31 2 0.517280558 0 NA
32 2 1.000000000 1 2
33 3 0.970310965 1 NA
34 3 0.920600257 1 NA
35 3 0.801496781 1 NA
36 3 0.635352677 0 NA
37 3 0.351879201 0 NA
38 3 0.133918706 0 NA
39 3 0.020005058 0 NA
40 3 -0.003554937 0 NA
41 3 -0.027281619 0 NA
42 3 -0.030216871 0 NA
43 3 -0.035568669 0 NA
44 3 -0.018927467 0 NA
45 3 -0.054635806 0 NA
46 3 -0.023042942 0 NA
47 3 -0.049607356 0 NA
48 3 1.000000000 1 4
>
CodePudding user response:
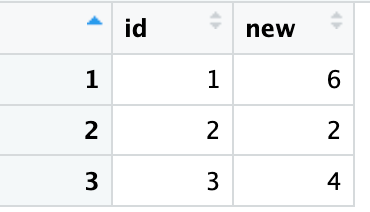
tmp %>%
dplyr::group_by(id)%>%
dplyr::summarise(new = sum(perc >= 0.8))
id new
<dbl> <int>
1 1 6
2 2 2
3 3 4
CodePudding user response:
I'm pretty not sure about your purpose, and please note that in id 2, perc is 10.9 then .635 which is smaller than 0.8, so maybe count you want is 1.(?)
Please check this out with dput object(tmp) you provided.
tmp %>%
group_by(id) %>%
mutate(key = (cumsum(perc < 0.8) == 0) * (perc > 0.8)) %>%
summarize(key = sum(key))
id key
<dbl> <int>
1 1 6
2 2 1
3 3 3