I've created a scatter plot (actually two similar subplots) using matplotlib.pyplot which I'm using for stylometric text analysis. The code I'm using to make the plot is as follows:
import matplotlib.pyplot as plt
import numpy as np
clusters = 4
two_d_matrix = np.array([[0.00617068, -0.53451777], [-0.01837677, -0.47131886], ...])
my_labels = [0, 1, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3]
fig, (plot1, plot2) = plt.subplots(1, 2, sharex=False, sharey=False, figsize=(20, 10))
plot1.axhline(0, color='#afafaf')
plot1.axvline(0, color='#afafaf')
for i in range(clusters):
try:
plot1.scatter(two_d_matrix[i:, 0], two_d_matrix[i:, 1], s=30, c=my_labels, cmap='viridis')
except (KeyError, ValueError) as e:
pass
plot1.legend(my_labels)
plot1.set_title("My First Plot")
plot2.axhline(0, color='#afafaf')
plot2.axvline(0, color='#afafaf')
for i in range(clusters):
try:
plot2.scatter(two_d_matrix[i:, 0], two_d_matrix[i:, 1], s=30, c=my_labels, cmap='viridis')
except (KeyError, ValueError) as e:
pass
plot2.legend(my_labels)
plot2.set_title("My Second Plot")
plt.show()
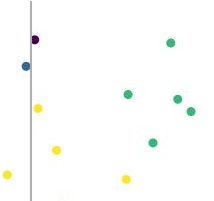
Because there are four distinct values in my_labels there are four colours which appear on the plot, these should correspond to the four clusters I expected to find.
The problem is that the legend only has three values, corresponding to the first three values in my_labels. It also appears that the legend isn't displaying a key for each colour, but for each of the axes and then for one of the colours. This means that the colours appearing in the plot are not matched to what appears in the legend, so the legend is inaccurate. I have no idea why this is happening.
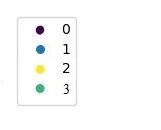
Ideally, the legend should display one colour for each unique value in my_labels, so it should look like this:
How can I get the legend to accurately display all the values it should be showing, i.e. one for each colour which appears in the plot?
CodePudding user response:
Before calling plot1.legend or plot2.legend, you can pass label = None to plot1.axhline or axvline (and similarly to plot2.axhline or plot2.axvline.) This will make sure it doesn't interfere with plotting legends of the scatter points and also not label those lines.
To get labels for all categories of scatter points, you'll have to call plot1.scatter or plot2.scatter by passing the label and choosing only values from two_d_matrix whose index matches with the index of label in my_labels.
You can do it as follows:
import matplotlib.pyplot as plt
import numpy as np
# Generate some (pseudo) random data which is reproducible
generator = np.random.default_rng(seed=121)
matrix = generator.uniform(size=(40, 2))
matrix = np.sort(matrix)
clusters = 4
my_labels = np.array([0, 1, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3])
fig, ax = plt.subplots(1, 1)
# Select data points wisely
for i in range(clusters):
pos = np.where(my_labels == i)
ax.scatter(matrix[pos, 0], matrix[pos, 1], s=30, cmap='viridis', label=i)
ax.axhline(0, color='#afafaf', label=None)
ax.axvline(0, color='#afafaf', label=None)
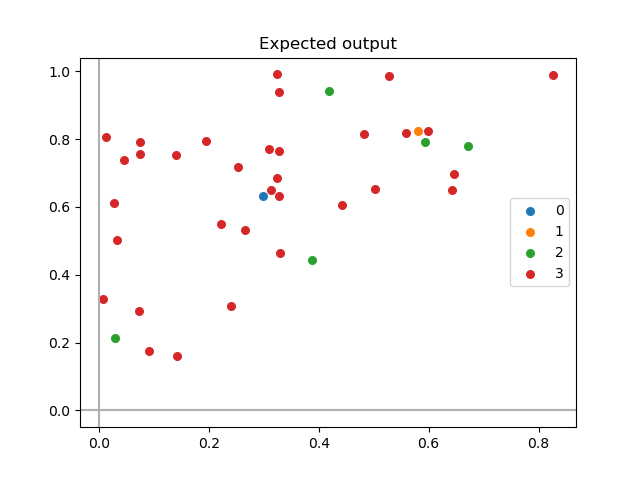
ax.legend()
ax.set_title("Expected output")
plt.show()
This gives:
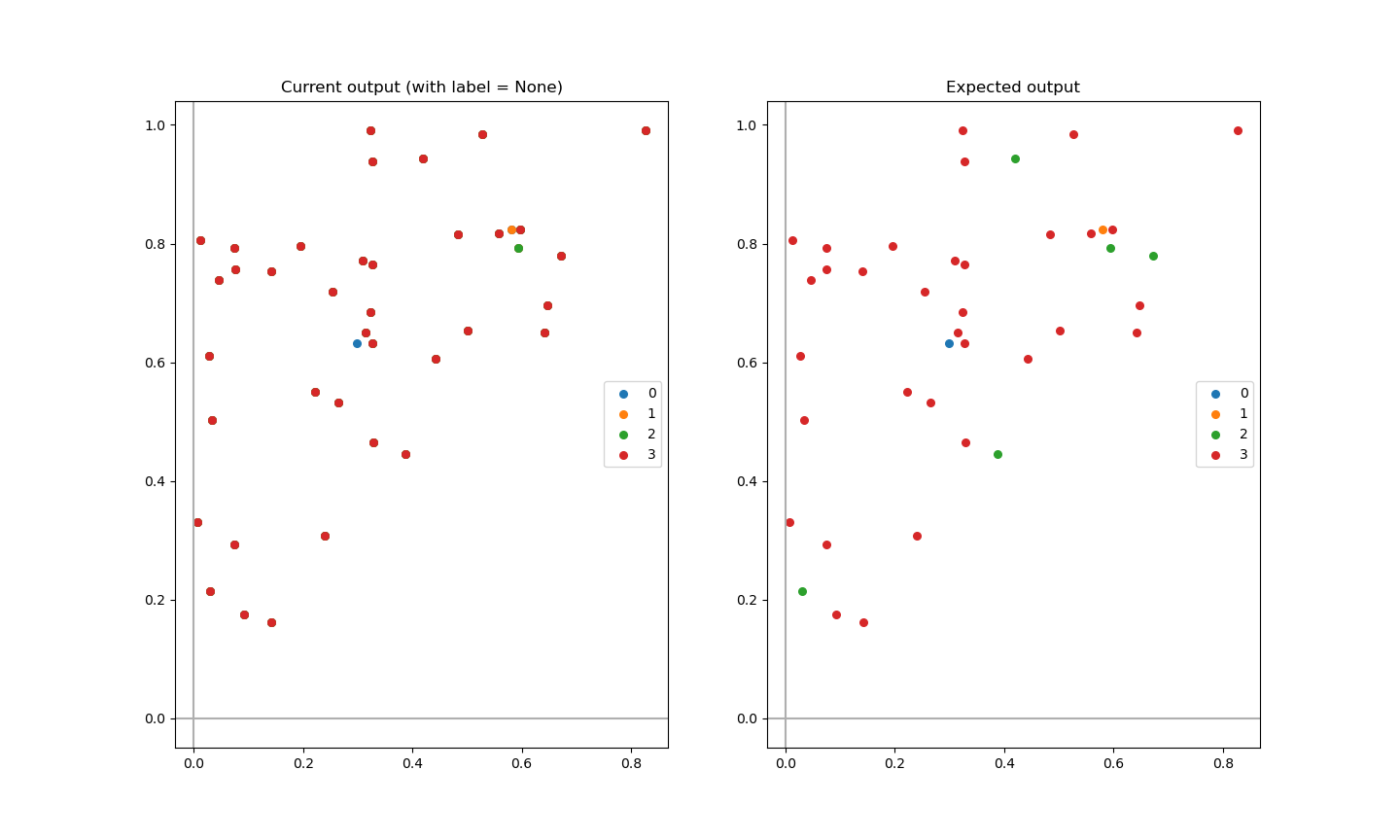
Comparison of current output and expected output
Observe how data points selection (done inside the for loops in the code below) affects the output:
Code:
import matplotlib.pyplot as plt
import numpy as np
# Generate some (pseudo) random data which is reproducible
generator = np.random.default_rng(seed=121)
matrix = generator.uniform(size=(40, 2))
matrix = np.sort(matrix)
clusters = 4
my_labels = np.array([0, 1, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3])
fig, ax = plt.subplots(1, 2)
# Question plot
for i in range(clusters):
ax[0].scatter(matrix[i:, 0], matrix[i:, 1], s=30, cmap='viridis', label=i)
ax[0].axhline(0, color='#afafaf', label=None)
ax[0].axvline(0, color='#afafaf', label=None)
ax[0].legend()
ax[0].set_title("Current output (with label = None)")
# Answer plot
for i in range(clusters):
pos = np.where(my_labels == i) # <- choose index of data points based on label position in my_labels
ax[1].scatter(matrix[pos, 0], matrix[pos, 1], s=30, cmap='viridis', label=i)
ax[1].axhline(0, color='#afafaf', label=None)
ax[1].axvline(0, color='#afafaf', label=None)
ax[1].legend()
ax[1].set_title("Expected output")
plt.show()