I have a forest inventory dataset of trees from 89 plots with different ground areas (0.16ha, 1ha, 2.5ha etc). To make the data suitable for my analysis, I have to extrapolate the population of the larger plots with reference to the smallest plot (0.16ha) and then randomly sample elements from the larger plots to make up the expected population (Population of the Large plot * area of smallest plot/area of the large plot). In doing this, each of the larger plots would have a different expected population.
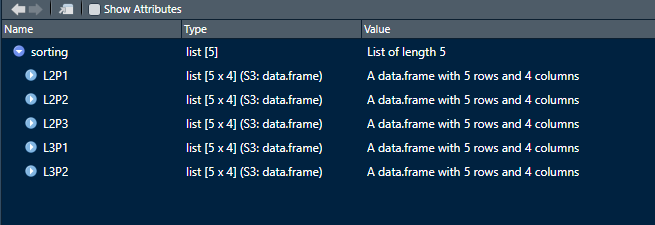
I can randomly select a single size from all plots in the list but I having difficulty modifying my code to randomly select a different size from the plots in the list. For example, in the data provided 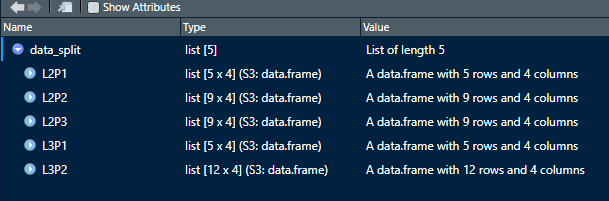 .
.
# Import tree data
species_data <- read.csv("stackoverflow_help.csv", header = TRUE, stringsAsFactors = FALSE)
# Split the dataset into a list of plots
data_split <- split( species_data , f = species_data$Plot)
# creating a loop to randomly select 5 individuals from the each plot (without replacing after selection)
library(dplyr)
fun2<- function (filename) {
sample_n(filename, size=5, replace=F)
}
# apply the loop
sorting <- lapply(data_split, fun2)
# result from the random sampling
How can the size=5 in the loop be modified to size = sizes <- c(4, 6, 8, 4, 10) and applied to each plot in the list? So 4, 6, 8, 4, and 10 elements will be sampled from L2P1, L2P2, L2P3, L3P1 and L3P2 respectively.
Thank you.
CodePudding user response:
If you want to iterate through multiple data objects as parameters passed to a function use Map instead of lapply:
fun2<- function (filename, size) {
sample_n(filename, size=size, replace=F)
}
sizes <- c(4, 6, 8, 4, 10)
sorting <- Map(fun2, filename = data_split, size = sizes)
An alternative is to use lapply with indices on your two-parameter function:
sorting <- lapply(seq_along(data_split), function(i) {
fun2(data_split[[i]], sizes[i]))
})
Both of which result in data like this:
sorting
#> $L2P1
#> Stand.ID Plot Plot_Name Species
#> 1 30977 L2P1 Aking plot 1, Oban forest Anonidium mannii
#> 2 30979 L2P1 Aking plot 1, Oban forest Caloncoba gilgiana
#> 3 30976 L2P1 Aking plot 1, Oban forest Anonidium mannii
#> 4 30978 L2P1 Aking plot 1, Oban forest Caloncoba gilgiana
#>
#> $L2P2
#> Stand.ID Plot Plot_Name Species
#> 1 31051 L2P2 Aking plot 2, Oban forest Cola rostrata
#> 2 31048 L2P2 Aking plot 2, Oban forest Angylocalyx oligophyllus
#> 3 31052 L2P2 Aking plot 2, Oban forest Cola verticillata
#> 4 31049 L2P2 Aking plot 2, Oban forest Anonidium mannii
#> 5 31054 L2P2 Aking plot 2, Oban forest Diospyros mespiliformis
#> 6 31056 L2P2 Aking plot 2, Oban forest Diospyros mespiliformis
#>
#> $L2P3
#> Stand.ID Plot Plot_Name Species
#> 1 31131 L2P3 Aking plot 3, Oban forest Angylocalyx oligophyllus
#> 2 31133 L2P3 Aking plot 3, Oban forest Caula edulis
#> 3 31138 L2P3 Aking plot 3, Oban forest Diospyros zenkeri
#> 4 31135 L2P3 Aking plot 3, Oban forest Dacryodes edulis
#> 5 31134 L2P3 Aking plot 3, Oban forest Coelocaryon preussii
#> 6 31132 L2P3 Aking plot 3, Oban forest Caula edulis
#> 7 31137 L2P3 Aking plot 3, Oban forest Diospyros mespiliformis
#> 8 31139 L2P3 Aking plot 3, Oban forest Diospyros zenkeri
#>
#> $L3P1
#> Stand.ID Plot Plot_Name Species
#> 1 31198 L3P1 Aking plot 4, Oban forest Carpolobia alba
#> 2 31199 L3P1 Aking plot 4, Oban forest Caula edulis
#> 3 31264 L3P1 Aking plot 4, Oban forest Trichilia obovoidea
#> 4 31200 L3P1 Aking plot 4, Oban forest Caula edulis
#>
#> $L3P2
#> Stand.ID Plot Plot_Name Species
#> 1 31276 L3P2 Aking plot 5, Oban forest Diospyros zenkeri
#> 2 31269 L3P2 Aking plot 5, Oban forest Calpocalyx cauliflorus
#> 3 31270 L3P2 Aking plot 5, Oban forest Coelocaryon preussii
#> 4 31273 L3P2 Aking plot 5, Oban forest Coelocaryon preussii
#> 5 31267 L3P2 Aking plot 5, Oban forest Angylocalyx oligophyllus
#> 6 31266 L3P2 Aking plot 5, Oban forest Angylocalyx oligophyllus
#> 7 31275 L3P2 Aking plot 5, Oban forest Cola lepidota
#> 8 31265 L3P2 Aking plot 5, Oban forest Angylocalyx oligophyllus
#> 9 31274 L3P2 Aking plot 5, Oban forest Cola lepidota
#> 10 31272 L3P2 Aking plot 5, Oban forest Coelocaryon preussii