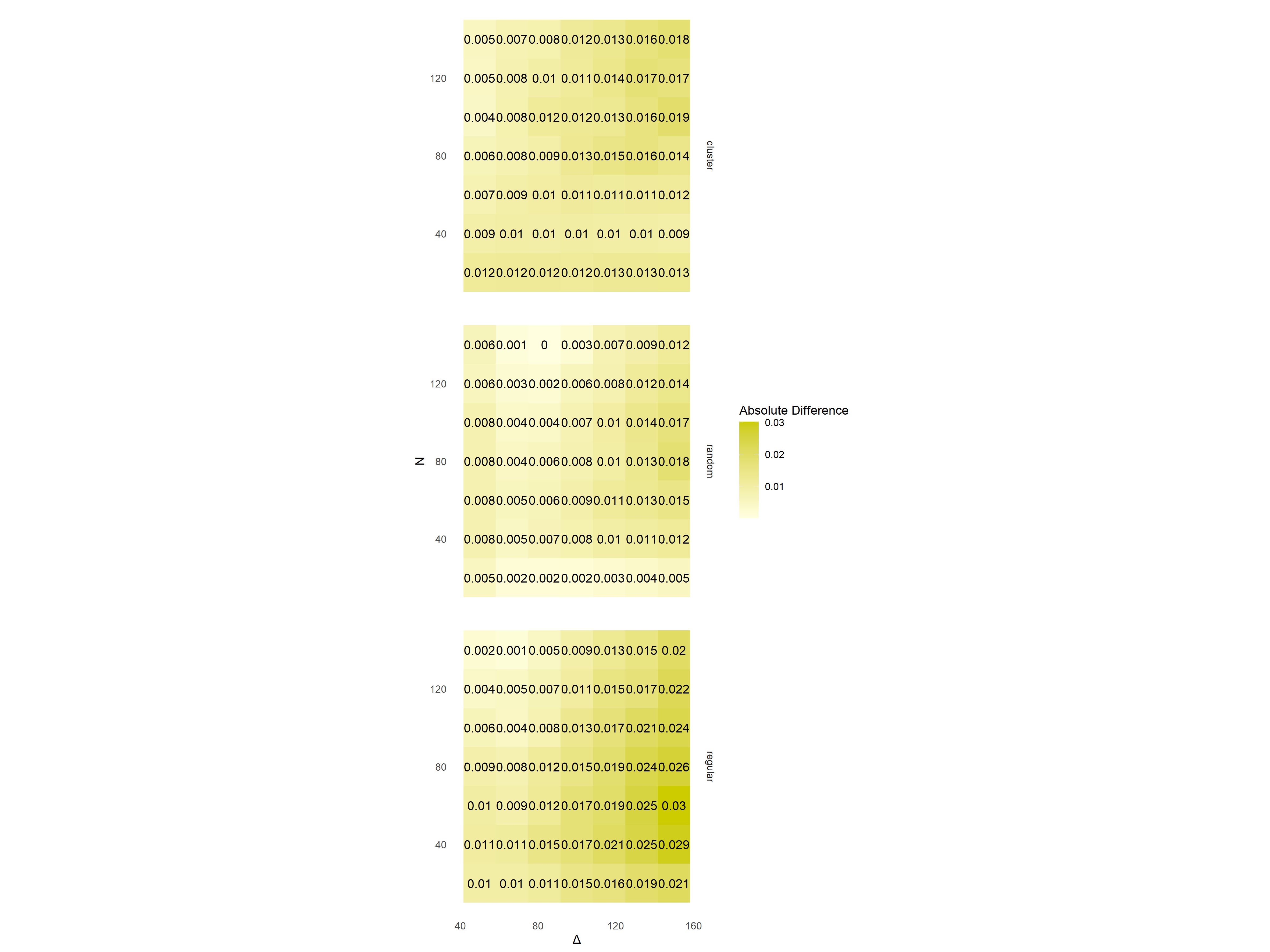
How do I assign a 1:1 aspect ratio to a facet heat map grid made with ggplot?
The aspect ratio to my facet grid is not 1:1, as can be seen below.
I have tried both assigning coord_equal and space = "free". This has worked previously however now this does not work after repeating the experiment. Is it possible that there is something wrong with the data input itself?
The code for generating these samples is this.
####input data for the figures, using dput
sum.q<-structure(list(beta = list(`1` = 50, `1` = 50, `1` = 50, `1` = 50,
`1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50,
`1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50,
`1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50,
`1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50,
`1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50,
`1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50,
`1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50,
`1` = 50, `1` = 50, `1` = 50, `2` = 50, `2` = 50, `2` = 50,
`2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50,
`2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50,
`2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50,
`2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50,
`2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50,
`2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50,
`2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50,
`2` = 50, `2` = 50, `2` = 50, `2` = 50, `3` = 150, `3` = 150,
`3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150,
`3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150,
`3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150,
`3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150,
`3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150,
`3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150,
`3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150,
`3` = 150, `3` = 150, `3` = 150, `3` = 150, `3` = 150, `4` = 150,
`4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150,
`4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150,
`4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150,
`4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150,
`4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150,
`4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150,
`4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150,
`4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150, `4` = 150),
theta = list(`1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20,
`1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20,
`1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20,
`1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20,
`1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20,
`1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20,
`1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20,
`1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20,
`1` = 20, `1` = 20, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `2` = 140, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `2` = 140, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `2` = 140, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `2` = 140, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `2` = 140, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `2` = 140, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `2` = 140, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `2` = 140, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `2` = 140, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20,
`3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20,
`3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20,
`3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20,
`3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20,
`3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20,
`3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20,
`3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20, `3` = 20,
`3` = 20, `3` = 20, `4` = 140, `4` = 140, `4` = 140,
`4` = 140, `4` = 140, `4` = 140, `4` = 140, `4` = 140,
`4` = 140, `4` = 140, `4` = 140, `4` = 140, `4` = 140,
`4` = 140, `4` = 140, `4` = 140, `4` = 140, `4` = 140,
`4` = 140, `4` = 140, `4` = 140, `4` = 140, `4` = 140,
`4` = 140, `4` = 140, `4` = 140, `4` = 140, `4` = 140,
`4` = 140, `4` = 140, `4` = 140, `4` = 140, `4` = 140,
`4` = 140, `4` = 140, `4` = 140, `4` = 140, `4` = 140,
`4` = 140, `4` = 140, `4` = 140, `4` = 140, `4` = 140,
`4` = 140, `4` = 140, `4` = 140, `4` = 140, `4` = 140,
`4` = 140), frequency = list(`1` = 15, `1` = 15, `1` = 15,
`1` = 15, `1` = 15, `1` = 15, `1` = 15, `1` = 32.5, `1` = 32.5,
`1` = 32.5, `1` = 32.5, `1` = 32.5, `1` = 32.5, `1` = 32.5,
`1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50,
`1` = 50, `1` = 67.5, `1` = 67.5, `1` = 67.5, `1` = 67.5,
`1` = 67.5, `1` = 67.5, `1` = 67.5, `1` = 85, `1` = 85,
`1` = 85, `1` = 85, `1` = 85, `1` = 85, `1` = 85, `1` = 102.5,
`1` = 102.5, `1` = 102.5, `1` = 102.5, `1` = 102.5, `1` = 102.5,
`1` = 102.5, `1` = 120, `1` = 120, `1` = 120, `1` = 120,
`1` = 120, `1` = 120, `1` = 120, `2` = 15, `2` = 15,
`2` = 15, `2` = 15, `2` = 15, `2` = 15, `2` = 15, `2` = 32.5,
`2` = 32.5, `2` = 32.5, `2` = 32.5, `2` = 32.5, `2` = 32.5,
`2` = 32.5, `2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50,
`2` = 50, `2` = 50, `2` = 67.5, `2` = 67.5, `2` = 67.5,
`2` = 67.5, `2` = 67.5, `2` = 67.5, `2` = 67.5, `2` = 85,
`2` = 85, `2` = 85, `2` = 85, `2` = 85, `2` = 85, `2` = 85,
`2` = 102.5, `2` = 102.5, `2` = 102.5, `2` = 102.5, `2` = 102.5,
`2` = 102.5, `2` = 102.5, `2` = 120, `2` = 120, `2` = 120,
`2` = 120, `2` = 120, `2` = 120, `2` = 120, `3` = 15,
`3` = 15, `3` = 15, `3` = 15, `3` = 15, `3` = 15, `3` = 15,
`3` = 32.5, `3` = 32.5, `3` = 32.5, `3` = 32.5, `3` = 32.5,
`3` = 32.5, `3` = 32.5, `3` = 50, `3` = 50, `3` = 50,
`3` = 50, `3` = 50, `3` = 50, `3` = 50, `3` = 67.5, `3` = 67.5,
`3` = 67.5, `3` = 67.5, `3` = 67.5, `3` = 67.5, `3` = 67.5,
`3` = 85, `3` = 85, `3` = 85, `3` = 85, `3` = 85, `3` = 85,
`3` = 85, `3` = 102.5, `3` = 102.5, `3` = 102.5, `3` = 102.5,
`3` = 102.5, `3` = 102.5, `3` = 102.5, `3` = 120, `3` = 120,
`3` = 120, `3` = 120, `3` = 120, `3` = 120, `3` = 120,
`4` = 15, `4` = 15, `4` = 15, `4` = 15, `4` = 15, `4` = 15,
`4` = 15, `4` = 32.5, `4` = 32.5, `4` = 32.5, `4` = 32.5,
`4` = 32.5, `4` = 32.5, `4` = 32.5, `4` = 50, `4` = 50,
`4` = 50, `4` = 50, `4` = 50, `4` = 50, `4` = 50, `4` = 67.5,
`4` = 67.5, `4` = 67.5, `4` = 67.5, `4` = 67.5, `4` = 67.5,
`4` = 67.5, `4` = 85, `4` = 85, `4` = 85, `4` = 85, `4` = 85,
`4` = 85, `4` = 85, `4` = 102.5, `4` = 102.5, `4` = 102.5,
`4` = 102.5, `4` = 102.5, `4` = 102.5, `4` = 102.5, `4` = 120,
`4` = 120, `4` = 120, `4` = 120, `4` = 120, `4` = 120,
`4` = 120), samplesize = list(`1` = 15, `1` = 32.5, `1` = 50,
`1` = 67.5, `1` = 85, `1` = 102.5, `1` = 120, `1` = 15,
`1` = 32.5, `1` = 50, `1` = 67.5, `1` = 85, `1` = 102.5,
`1` = 120, `1` = 15, `1` = 32.5, `1` = 50, `1` = 67.5,
`1` = 85, `1` = 102.5, `1` = 120, `1` = 15, `1` = 32.5,
`1` = 50, `1` = 67.5, `1` = 85, `1` = 102.5, `1` = 120,
`1` = 15, `1` = 32.5, `1` = 50, `1` = 67.5, `1` = 85,
`1` = 102.5, `1` = 120, `1` = 15, `1` = 32.5, `1` = 50,
`1` = 67.5, `1` = 85, `1` = 102.5, `1` = 120, `1` = 15,
`1` = 32.5, `1` = 50, `1` = 67.5, `1` = 85, `1` = 102.5,
`1` = 120, `2` = 15, `2` = 32.5, `2` = 50, `2` = 67.5,
`2` = 85, `2` = 102.5, `2` = 120, `2` = 15, `2` = 32.5,
`2` = 50, `2` = 67.5, `2` = 85, `2` = 102.5, `2` = 120,
`2` = 15, `2` = 32.5, `2` = 50, `2` = 67.5, `2` = 85,
`2` = 102.5, `2` = 120, `2` = 15, `2` = 32.5, `2` = 50,
`2` = 67.5, `2` = 85, `2` = 102.5, `2` = 120, `2` = 15,
`2` = 32.5, `2` = 50, `2` = 67.5, `2` = 85, `2` = 102.5,
`2` = 120, `2` = 15, `2` = 32.5, `2` = 50, `2` = 67.5,
`2` = 85, `2` = 102.5, `2` = 120, `2` = 15, `2` = 32.5,
`2` = 50, `2` = 67.5, `2` = 85, `2` = 102.5, `2` = 120,
`3` = 15, `3` = 32.5, `3` = 50, `3` = 67.5, `3` = 85,
`3` = 102.5, `3` = 120, `3` = 15, `3` = 32.5, `3` = 50,
`3` = 67.5, `3` = 85, `3` = 102.5, `3` = 120, `3` = 15,
`3` = 32.5, `3` = 50, `3` = 67.5, `3` = 85, `3` = 102.5,
`3` = 120, `3` = 15, `3` = 32.5, `3` = 50, `3` = 67.5,
`3` = 85, `3` = 102.5, `3` = 120, `3` = 15, `3` = 32.5,
`3` = 50, `3` = 67.5, `3` = 85, `3` = 102.5, `3` = 120,
`3` = 15, `3` = 32.5, `3` = 50, `3` = 67.5, `3` = 85,
`3` = 102.5, `3` = 120, `3` = 15, `3` = 32.5, `3` = 50,
`3` = 67.5, `3` = 85, `3` = 102.5, `3` = 120, `4` = 15,
`4` = 32.5, `4` = 50, `4` = 67.5, `4` = 85, `4` = 102.5,
`4` = 120, `4` = 15, `4` = 32.5, `4` = 50, `4` = 67.5,
`4` = 85, `4` = 102.5, `4` = 120, `4` = 15, `4` = 32.5,
`4` = 50, `4` = 67.5, `4` = 85, `4` = 102.5, `4` = 120,
`4` = 15, `4` = 32.5, `4` = 50, `4` = 67.5, `4` = 85,
`4` = 102.5, `4` = 120, `4` = 15, `4` = 32.5, `4` = 50,
`4` = 67.5, `4` = 85, `4` = 102.5, `4` = 120, `4` = 15,
`4` = 32.5, `4` = 50, `4` = 67.5, `4` = 85, `4` = 102.5,
`4` = 120, `4` = 15, `4` = 32.5, `4` = 50, `4` = 67.5,
`4` = 85, `4` = 102.5, `4` = 120), absdif = c(0.0119600832573928,
0.0061853720011284, 0.00403148727084383, 0.00171816611895332,
0.00475096240489938, 0.00361294992989708, 0.00283390673322197,
0.0197653803402051, 0.0116289721462817, 0.00691163964232991,
0.0034994514497768, 0.000729115887676227, 0.00591051420356356,
0.00432901651763746, 0.00885174637334012, 0.0105524824243402,
0.0110461943685039, 0.00649988373294956, 0.00408903822169815,
0.000701342665712215, 0.00674447913502476, 0.00656095632233141,
0.0014860229861838, 0.00756139082735856, 0.0114628610351705,
0.00782035411305263, 0.00538158452407454, 0.000447198266378379,
0.0103915829030034, 0.00381636726565354, 0.00270657601626321,
0.00615261421482005, 0.0116139721462817, 0.00710764219534272,
0.00442954556233189, 0.101865558508852, 0.0156045360852814,
0.00569478656427126, 0.00325694517066357, 0.00595385673032028,
0.0116284165907261, 0.00713296662400191, 0.0490494135182385,
0.0408816244850832, 0.0195526442621871, 0.0106228320701901,
0.00777291395429778, 0.00315277795326487, 0.0139250832573928,
0.00676089859503747, 0.00250609152779393, 0.000880309072746346,
0.00292483331645765, 0.00160205400043145, 0.000947636067078293,
9.17954469776441e-05, 0.0215851803910177, 0.00688534303948191,
0.0037449816794209, 0.00188897340893446, 0.0043147703958005,
0.0022989841073696, 0.00130670532938834, 0.0206652859934779,
0.0166789742421427, 0.00613867637281524, 0.00348146376888738,
0.00299593282237846, 0.00519485091146531, 0.0028047262664508,
0.0125823090626575, 0.00897393460619266, 0.0144965568419247,
0.00730312081725969, 0.00487236826595917, 0.00272364593204483,
0.00696185238783898, 0.0234819438903975, 0.00876896310194339,
0.00731230696778485, 0.0131225739784452, 0.00735812081725969,
0.00298975252825848, 0.00316313716250295, 0.0603461244810728,
0.0219946716373404, 0.00852673709150694, 0.00578475374270784,
0.014615002359405, 0.00823867637281524, 0.00361289971405382,
0.0417237974954581, 0.0580745381811425, 0.0278557121836455,
0.0172100686008853, 0.00215442789295837, 0.0205594199652945,
0.0116753430394819, 0.0109929205324424, 0.00659264416158682,
0.0123665805327734, 0.0103911873421071, 0.00878293368873167,
0.007128263866953, 0.00818528818495187, 0.0130749099188775,
0.0108318094213313, 0.0043432192312718, 0.0139006573708434,
0.0111770429268607, 0.0102919151823733, 0.00773731356858547,
0.0237869441913092, 0.0138842029667243, 0.0100340316435535,
0.00373999817428886, 0.0143930228311461, 0.0120550182848421,
0.010292837258888, 0.0467935249800841, 0.0224700874491149,
0.0144173797699672, 0.0106862538657757, 0.00377401846756686,
0.014774187619367, 0.0121371026819308, 0.0136025243902338,
0.0158081645658228, 0.022497474870901, 0.0136208023460143,
0.0099918094213313, 0.00321460635717812, 0.0153486700850976,
0.00942348923020696, 0.00414134766214742, 0.00840322310821114,
0.0199911758106088, 0.014023109743689, 0.0100256983102202,
0.00279263851972684, 0.0205673327258088, 0.0129185315645828,
0.0121924537223062, 0.000344671048373407, 0.0243132547947506,
0.0181324883280655, 0.0165668094213313, 0.0174306364241788,
0.0140661922726428, 0.00742571402295568, 0.00594521690844399,
0.00367027570884202, 0.00610839971788404, 0.00481830676882413,
0.0476125315239956, 0.0186839697575122, 0.0201224985617497,
0.0116205355375283, 0.00818570817743713, 0.00416929393395373,
0.0077076614377661, 0.0689361434649397, 0.0406036384259798,
0.01799174753529, 0.0204264814252292, 0.0138981429644011,
0.0103300831865573, 0.00433475591748193, 0.101426485769379,
0.0545986184495223, 0.035571970496575, 0.0184189697575122,
0.0252984974887139, 0.0192871635734258, 0.0133717469661655,
0.0301115693666431, 0.0758834228783093, 0.046438860944897,
0.0376921550737752, 0.0202878586464011, 0.0280479687717132,
0.0195289235418552, 0.0859120866013194, 0.0355070499189914,
0.069510059566544, 0.0505862616525466, 0.0356901394655923,
0.0199606364241788, 0.0291304877823563, 0.1615021887603,
0.121179287777907, 0.0754947496402514, 0.0912408887538834,
0.0764375431145627, 0.0589816440921819, 0.0382239697575122
)), class = c("grouped_df", "tbl_df", "tbl", "data.frame"
), row.names = c(NA, -196L), groups = structure(list(beta = list(
`1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50, `1` = 50,
`1` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50, `2` = 50,
`2` = 50, `2` = 50, `3` = 150, `3` = 150, `3` = 150, `3` = 150,
`3` = 150, `3` = 150, `3` = 150, `4` = 150, `4` = 150, `4` = 150,
`4` = 150, `4` = 150, `4` = 150, `4` = 150), theta = list(
`1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20, `1` = 20,
`1` = 20, `2` = 140, `2` = 140, `2` = 140, `2` = 140, `2` = 140,
`2` = 140, `2` = 140, `3` = 20, `3` = 20, `3` = 20, `3` = 20,
`3` = 20, `3` = 20, `3` = 20, `4` = 140, `4` = 140, `4` = 140,
`4` = 140, `4` = 140, `4` = 140, `4` = 140), frequency = list(
`1` = 15, `1` = 32.5, `1` = 50, `1` = 67.5, `1` = 85, `1` = 102.5,
`1` = 120, `2` = 15, `2` = 32.5, `2` = 50, `2` = 67.5, `2` = 85,
`2` = 102.5, `2` = 120, `3` = 15, `3` = 32.5, `3` = 50, `3` = 67.5,
`3` = 85, `3` = 102.5, `3` = 120, `4` = 15, `4` = 32.5, `4` = 50,
`4` = 67.5, `4` = 85, `4` = 102.5, `4` = 120), .rows = structure(list(
1:7, 8:14, 15:21, 22:28, 29:35, 36:42, 43:49, 50:56, 57:63,
64:70, 71:77, 78:84, 85:91, 92:98, 99:105, 106:112, 113:119,
120:126, 127:133, 134:140, 141:147, 148:154, 155:161, 162:168,
169:175, 176:182, 183:189, 190:196), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -28L), .drop = TRUE))
#####generate the figures
reldifgg<-ggplot(sum.qmerge,aes(x=as.numeric(beta),y=as.numeric(theta), fill=absdif))
geom_tile()
scale_fill_gradient(low="lightyellow",
high="#CCCC00",
name="Absolute Difference")
theme_minimal()
ylab("N")
xlab("\U0394")
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank()),aspect.ratio = 1)
geom_text(aes(label = round(absdif, 3)),colour="black")
plotforresultsbetatheta<-reldifgg facet_grid((sum.qmerge$landscape)~.,space = "free")
coord_equal()
####save the file
ggsave(plotforresultsbetatheta,width=40,height=30, units="cm", file="plotforresultsbetathetaabsdif1.png")
CodePudding user response:
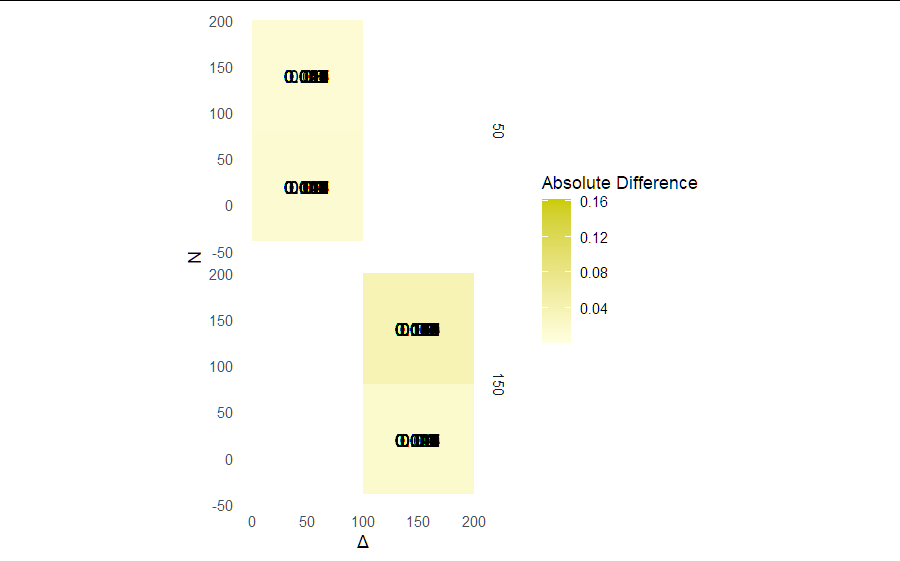
Attention: The dataset is adapted. In my point of view there are some issues with your code:
- sum.qmerge is not provided. I guess it is sum.q
- I have adapted the code a little and added
theme(aspect.ratio = 1)after removing,space = "free". - unfortunately there is no
landscapecolumn so I have usedbetafor demonstration purposes.
library(tidyverse)
df <- sum.q %>%
unnest()
ggplot(df,aes(x=as.numeric(beta),y=as.numeric(theta), fill=absdif))
geom_tile()
scale_fill_gradient(low="lightyellow",
high="#CCCC00",
name="Absolute Difference")
theme_minimal()
ylab("N")
xlab("\U0394")
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
geom_text(aes(label = round(absdif, 3)),colour="black")
facet_grid(beta ~.)
theme(aspect.ratio = 1)