I wrote some R code for simulating random samples from a Poisson distribution, based on the description of an algorithm (see attached image). But my code does not seem to work correctly, because the generated random samples are of a different pattern compared with those generated by R's built-in rpois() function. Can anybody tell me what I did wrong and how to fix my function?
r.poisson <- function(n, l=0.5)
{
U <- runif(n)
X <- rep(0,n)
p=exp(-l)
F=p
for(i in 1:n)
{
if(U[i] < F)
{
X[i] <- i
} else
{
p=p*l/(i 1)
F=F p
i=i 1
}
}
return(X)
}
r.poisson(50)
The output is very different from rpois(50, lambda = 0.5). The algorithm I followed is:

CodePudding user response:
(Thank you for your question. Now I know how a Poisson random variable is simulated.)
You had a misunderstanding. The inverse CDF method (with recursive computation) you referenced is used to generate a single Poisson random sample. So you need to fix this function to produce a single number. Here is the correct function, commented to help you follow each step.
rpois1 <- function (lambda) {
## step 1
U <- runif(1)
## step 2
i <- 0
p <- exp(-lambda)
F <- p
## you need an "infinite" loop
## no worry, it will "break" at some time
repeat {
## step 3
if (U < F) {
X <- i
break
}
## step 4
i <- i 1
p <- lambda * p * i
F <- F p
## back to step 3
}
return(X)
}
Now to get n samples, you need to call this function n times. R has a nice function called replicate to repeat a function many times.
r.poisson <- function (n, lambda) {
## use `replicate()` to call `rpois1` n times
replicate(n, rpois1(lambda))
}
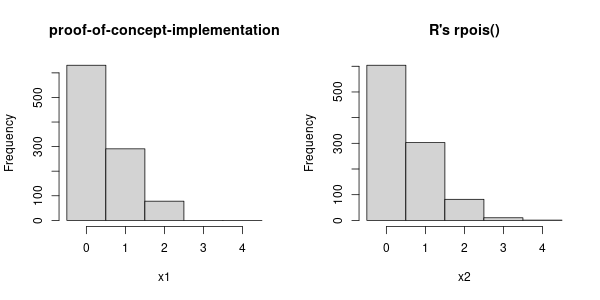
Now we can make a reasonable comparison with R's own rpois.
x1 <- r.poisson(1000, lambda = 0.5)
x2 <- rpois(1000, lambda = 0.5)
## set breaks reasonably when making a histogram
xmax <- max(x1, x2) 0.5
par(mfrow = c(1, 2))
hist(x1, main = "proof-of-concept-implementation", breaks = seq.int(-0.5, xmax))
hist(x2, main = "R's rpois()", breaks = seq.int(-0.5, xmax))
CodePudding user response:
A vectorized version will run much faster than a non-vectorized function using replicate. The idea is to iteratively drop the uniform random samples as i is incremented.
r.poisson1 <- function(n, l = 0.5) {
U <- runif(n)
i <- 0L
X <- integer(n)
p <- exp(-l)
F <- p
idx <- 1:n
while (length(idx)) {
bln <- U < F
X[idx[bln]] <- i
p <- l*p/(i <- i 1L)
F <- F p
idx <- idx[!bln]
U <- U[!bln]
}
X
}
@Zheyuan Li's non-vectorized functions:
rpois1 <- function (lambda) {
## step 1
U <- runif(1)
## step 2
i <- 0
p <- exp(-lambda)
F <- p
## you need an "infinite" loop
## no worry, it will "break" at some time
repeat {
## step 3
if (U < F) {
X <- i
break
}
## step 4
i <- i 1
p <- lambda * p * i
F <- F p
## back to step 3
}
return(X)
}
r.poisson2 <- function (n, lambda) {
## use `replicate()` to call `rpois1` n times
replicate(n, rpois1(lambda))
}
Benchmark:
microbenchmark::microbenchmark(r.poisson1(1e5),
r.poisson2(1e5, 0.5),
rpois(1e5, 0.5))
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> r.poisson1(1e 05) 3.063202 3.129151 3.782200 3.225402 3.734600 18.377700 100
#> r.poisson2(1e 05, 0.5) 217.631002 244.816601 269.692648 267.977001 287.599251 375.910601 100
#> rpois(1e 05, 0.5) 1.519901 1.552300 1.649026 1.579551 1.620451 7.531401 100