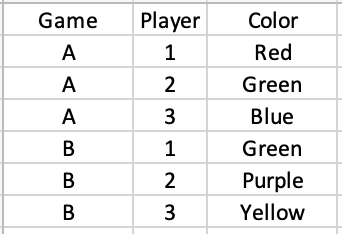
I am working with a data frame like the following, where Color and `Player are factor variables:
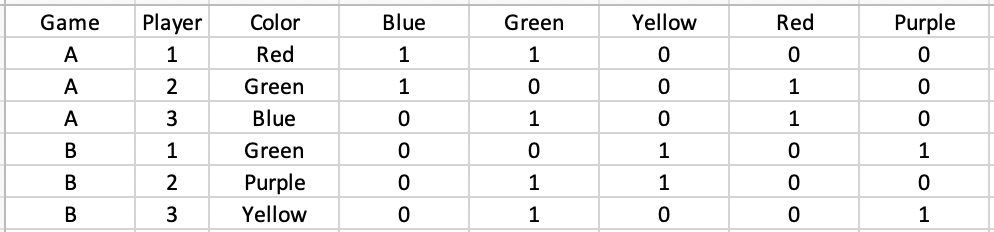
I want to create indicator variables for each value of the color column. However, I want those indicator variables to represent whether the color is present for other players in the same game (not whether it's present for that player). So I want the above table to turn into:
I imagine the code will have group_by(Game) %>%, but I'm lost beyond that.
Data:
structure(list(Game = c("A", "A", "A", "B", "B", "B"), Player = c(1L,
2L, 3L, 1L, 2L, 3L), Color = c("Red", "Green", "Blue", "Green",
"Purple", "Yellow"), Blue = c(1L, 1L, 0L, 0L, 0L, 0L), Green = c(1L,
0L, 1L, 0L, 1L, 1L), Yellow = c(0L, 0L, 0L, 1L, 1L, 0L), Red = c(0L,
1L, 1L, 0L, 0L, 0L), Purple = c(0L, 0L, 0L, 1L, 0L, 1L)), class = "data.frame", row.names = c(NA,
-6L))
CodePudding user response:
Perhaps this helps - split the 'Color' column by 'Game', create a binary matrix by comparing the elements of 'Color' (!=), convert to tibble, row bind (_dfr) and bind the dataset with the original dataset (bind_cols)
library(purrr)
library(dplyr)
library(tidyr)
map_dfr(split(df1$Color, df1$Game), ~ {
m1 <- (outer(.x, .x, FUN = `!=`))
colnames(m1) <- .x
as_tibble(m1)}) %>%
mutate(across(everything(), replace_na, 0)) %>%
bind_cols(df1, .)
-output
Game Player Color Red Green Blue Purple Yellow
1 A 1 Red 0 1 1 0 0
2 A 2 Green 1 0 1 0 0
3 A 3 Blue 1 1 0 0 0
4 B 1 Green 0 0 0 1 1
5 B 2 Purple 0 1 0 0 1
6 B 3 Yellow 0 1 0 1 0
Or another option is with dummy_cols and then modify the output
library(fastDummies)
library(stringr)
dummy_cols(df1, 'Color') %>%
rename_with(~ str_remove(.x, "Color_")) %>%
group_by(Game) %>%
mutate(across(Blue:Yellow, ~ (Color != cur_column() & any(.x)))) %>%
ungroup
-output
# A tibble: 6 × 8
Game Player Color Blue Green Purple Red Yellow
<chr> <int> <chr> <int> <int> <int> <int> <int>
1 A 1 Red 1 1 0 0 0
2 A 2 Green 1 0 0 1 0
3 A 3 Blue 0 1 0 1 0
4 B 1 Green 0 0 1 0 1
5 B 2 Purple 0 1 0 0 1
6 B 3 Yellow 0 1 1 0 0
data
df1 <- structure(list(Game = c("A", "A", "A", "B", "B", "B"), Player = c(1L,
2L, 3L, 1L, 2L, 3L), Color = c("Red", "Green", "Blue", "Green",
"Purple", "Yellow")), row.names = c(NA, -6L), class = "data.frame")
CodePudding user response:
Here is a way how we could do it:
First we use model.matrix() fucntion multiply it by 1 and substract 1 within a wrap of abs().
Then we get almost the desired output, the only thing that is left is the get zeros in case if non of the colors is present. We do this with a mutate across...:
library(dplyr)
df %>%
cbind(abs((model.matrix(~ Color 0, .) == 1)*1-1)) %>%
group_by(Game) %>%
mutate(across(-c(Player, Color), ~case_when(sum(.)==3 ~0,
TRUE ~ .)))
Game Player Color ColorBlue ColorGreen ColorPurple ColorRed ColorYellow
<chr> <int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 A 1 Red 1 1 0 0 0
2 A 2 Green 1 0 0 1 0
3 A 3 Blue 0 1 0 1 0
4 B 1 Green 0 0 1 0 1
5 B 2 Purple 0 1 0 0 1
6 B 3 Yellow 0 1 1 0 0
>
CodePudding user response:
Here is another approach using full_join and pivot_wider from tidyverse. I believe this also gives the same result. The filter is included to avoid same color indicators as 1.
library(tidyverse)
full_join(df, df, by = "Game", suffix = c("", "_Two")) %>%
filter(Color != Color_Two) %>%
mutate(val = 1) %>%
pivot_wider(id_cols = c(Game, Player, Color),
names_from = Color_Two,
values_from = val,
values_fill = 0)
Output
Game Player Color Green Blue Red Purple Yellow
<chr> <int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 A 1 Red 1 1 0 0 0
2 A 2 Green 0 1 1 0 0
3 A 3 Blue 1 0 1 0 0
4 B 1 Green 0 0 0 1 1
5 B 2 Purple 1 0 0 0 1
6 B 3 Yellow 1 0 0 1 0
CodePudding user response:
Another possible solution:
library(tidyverse)
df %>%
group_by(Game) %>%
mutate(aux = list(Color)) %>%
unnest(aux) %>%
filter(aux != Color) %>%
ungroup %>%
pivot_wider(Game:Color, names_from = aux, values_from = aux, values_fill = 0,
values_fn = length)
#> # A tibble: 6 × 8
#> Game Player Color Green Blue Red Purple Yellow
#> <chr> <int> <chr> <int> <int> <int> <int> <int>
#> 1 A 1 Red 1 1 0 0 0
#> 2 A 2 Green 0 1 1 0 0
#> 3 A 3 Blue 1 0 1 0 0
#> 4 B 1 Green 0 0 0 1 1
#> 5 B 2 Purple 1 0 0 0 1
#> 6 B 3 Yellow 1 0 0 1 0
CodePudding user response:
Using base R, you can write a small function and evaluate using tapply:
fun <- function(x) {
nms <- levels(x)
tab <- tcrossprod(table(x))
dimnames(tab) <- list(nms, nms)
tab[x, ]
}
data.frame(df1, do.call(rbind, with(df1, tapply(factor(Color), Game, fun))), row.names = NULL)
Game Player Color Blue Green Purple Red Yellow
1 A 1 Red 1 1 0 1 0
2 A 2 Green 1 1 0 1 0
3 A 3 Blue 1 1 0 1 0
4 B 1 Green 0 1 1 0 1
5 B 2 Purple 0 1 1 0 1
6 B 3 Yellow 0 1 1 0 1
Note that out of all the options given, This is by far the fastest, yet only using base R:
Here is the benchmark:
library(microbenchmark)
microbenchmark(Tarjae(df1), akrun(df1), ben(df1), onyambu(df1),
paulS(df1), unit = 'relative')
Unit: relative
expr min lq mean median uq max neval
Tarjae(df1) 18.775201 18.11495 13.533556 17.171485 15.746554 1.105045 100
akrun(df1) 9.755032 8.83519 7.137294 8.756033 8.241494 1.455906 100
ben(df1) 21.084371 18.57861 14.699821 17.950987 16.486863 3.124906 100
onyambu(df1) 1.000000 1.00000 1.000000 1.000000 1.000000 1.000000 100
paulS(df1) 33.108208 31.27110 24.918541 30.266024 27.420363 3.156215 100
For larger dataframes, some of the given code breaks down, while those that dont break down are still slow to the base R approach:
df2<- transform(data.frame(Game = sample(LETTERS, 2000, TRUE), Color = sample(colors(), 2000, TRUE)), Player = ave(Game, Game, FUN=seq_along))
microbenchmark(Tarjae(df2), akrun(df2), onyambu(df2), paulS(df2))
Unit: milliseconds
expr min lq mean median uq max neval
Tarjae(df2) 2147.67826 2234.5575 2460.1924 2423.20994 2653.1737 3049.9455 100
akrun(df2) 108.25249 121.3167 144.6715 130.48052 152.9518 404.7286 100
onyambu(df2) 67.19992 80.3653 111.2821 91.05784 118.4877 331.6724 100
paulS(df2) 183.88836 200.6224 231.0155 215.18942 237.5717 467.1721 100
Code for the benchmark:
Tarjae <- function(df){
df %>%
cbind(abs((model.matrix(~ Color 0, .) == 1)*1-1)) %>%
group_by(Game) %>%
mutate(across(-c(Player, Color), ~case_when(sum(.)==3 ~0,
TRUE ~ .)))
}
akrun <- function(df1){
map_dfr(split(df1$Color, df1$Game), ~ {
m1 <- (outer(.x, .x, FUN = `!=`))
colnames(m1) <- .x
as_tibble(m1)}) %>%
mutate(across(everything(), replace_na, 0)) %>%
bind_cols(df1, .)
}
ben <- function(df){
full_join(df, df, by = "Game", suffix = c("", "_Two")) %>%
filter(Color != Color_Two) %>%
mutate(val = 1) %>%
pivot_wider(id_cols = c(Game, Player, Color),
names_from = Color_Two,
values_from = val,
values_fill = 0)
}
onyambu <- function(df1){
fun <- function(x) {
nms <- levels(x)
tab <- tcrossprod(table(x))
dimnames(tab) <- list(nms, nms)
tab[x, ]
}
data.frame(df1, do.call(rbind, with(df1, tapply(factor(Color), Game, fun))), row.names = NULL)
}
paulS <- function(df){
df %>%
group_by(Game) %>%
mutate(aux = list(Color)) %>%
unnest(aux) %>%
filter(aux != Color) %>%
ungroup %>%
pivot_wider(Game:Color, names_from = aux, values_from = aux, values_fill = 0,
values_fn = length)
}