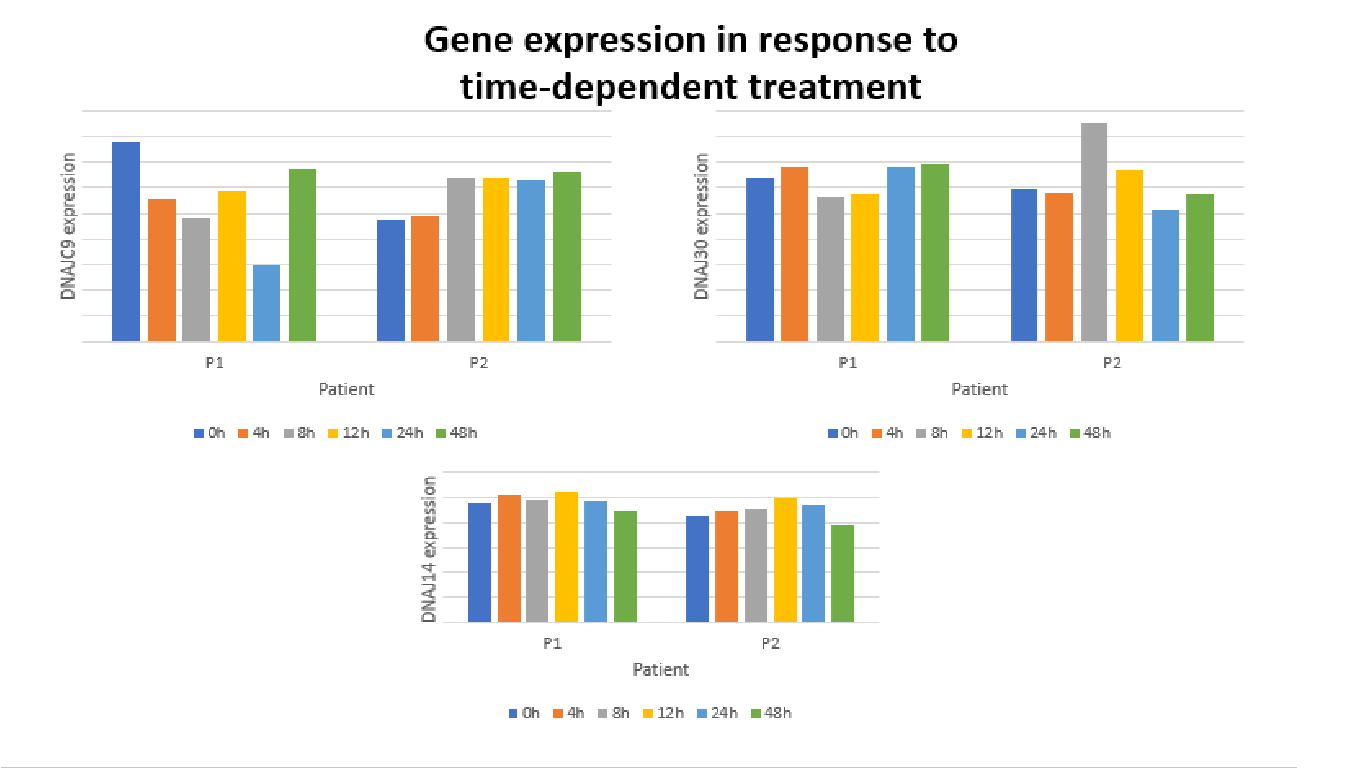
I want to create subplots a group Bar Charts for each gene, where the Y-axis are the Gene Expression values and the X-axis are the time points for each patient ID.
Column ID explanation:
0h_P1_T1_TimeC1_PIDC4_Non-Survivor
Time point: substring before the first substring (e.g., 0h) Patient ID: substring after the first substring (e.g., P1)
Code:
import pandas as pd
import plotly.graph_objects as go
import numpy as np
for exp in treatment_df:
for h in treatment_df.columns.str.split('_')[0][0]: # Get the "hours", which is the substring before the first underscore
fig = go.Figure(data=[go.Bar(name=h, x=treatment_df.index, y=exp)])
fig.show()
Traceback:
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-33-82ce32460b0d> in <module>()
2 for exp in treatment_df:
3 for h in treatment_df.columns.str.split('_')[0][0]: # Get the "hours", which is the substring before the first underscore
----> 4 fig = go.Figure(data=[go.Bar(name=h, x=treatment_df.index, y=exp)])
5 fig.show()
5 frames
/usr/local/lib/python3.7/dist-packages/plotly/graph_objs/_bar.py in __init__(self, arg, alignmentgroup, base, basesrc, cliponaxis, constraintext, customdata, customdatasrc, dx, dy, error_x, error_y, hoverinfo, hoverinfosrc, hoverlabel, hovertemplate, hovertemplatesrc, hovertext, hovertextsrc, ids, idssrc, insidetextanchor, insidetextfont, legendgroup, legendgrouptitle, legendrank, marker, meta, metasrc, name, offset, offsetgroup, offsetsrc, opacity, orientation, outsidetextfont, selected, selectedpoints, showlegend, stream, text, textangle, textfont, textposition, textpositionsrc, textsrc, texttemplate, texttemplatesrc, uid, uirevision, unselected, visible, width, widthsrc, x, x0, xaxis, xcalendar, xhoverformat, xperiod, xperiod0, xperiodalignment, xsrc, y, y0, yaxis, ycalendar, yhoverformat, yperiod, yperiod0, yperiodalignment, ysrc, **kwargs)
3208 _v = y if y is not None else _v
3209 if _v is not None:
-> 3210 self["y"] = _v
3211 _v = arg.pop("y0", None)
3212 _v = y0 if y0 is not None else _v
/usr/local/lib/python3.7/dist-packages/plotly/basedatatypes.py in __setitem__(self, prop, value)
4825 # ### Handle simple property ###
4826 else:
-> 4827 self._set_prop(prop, value)
4828 else:
4829 # Make sure properties dict is initialized
/usr/local/lib/python3.7/dist-packages/plotly/basedatatypes.py in _set_prop(self, prop, val)
5169 return
5170 else:
-> 5171 raise err
5172
5173 # val is None
/usr/local/lib/python3.7/dist-packages/plotly/basedatatypes.py in _set_prop(self, prop, val)
5164
5165 try:
-> 5166 val = validator.validate_coerce(val)
5167 except ValueError as err:
5168 if self._skip_invalid:
/usr/local/lib/python3.7/dist-packages/_plotly_utils/basevalidators.py in validate_coerce(self, v)
403 v = to_scalar_or_list(v)
404 else:
--> 405 self.raise_invalid_val(v)
406 return v
407
/usr/local/lib/python3.7/dist-packages/_plotly_utils/basevalidators.py in raise_invalid_val(self, v, inds)
297 typ=type_str(v),
298 v=repr(v),
--> 299 valid_clr_desc=self.description(),
300 )
301 )
ValueError:
Invalid value of type 'builtins.str' received for the 'y' property of bar
Received value: '0h_P1_T1_TimeC1_PIDC4_Non-Survivor'
The 'y' property is an array that may be specified as a tuple,
list, numpy array, or pandas Series
Data:
treatment_df.iloc[0:3:,0:12].head().to_dict()
{'0h_P1_T1_TimeC1_PIDC4_Non-Survivor': {'DNAJC14': 0.23768844221105523,
'DNAJC30': 0.12713567839195977,
'DNAJC9': 0.15527638190954773},
'0h_P2_T1_TimeC2_PIDC2_Survivor': {'DNAJC14': 0.2128966223132037,
'DNAJC30': 0.11873080859774823,
'DNAJC9': 0.09518935516888441},
'12h_P1_T4_TimeC2_PIDC4_Non-Survivor': {'DNAJC14': 0.26175869120654405,
'DNAJC30': 0.114519427402863,
'DNAJC9': 0.11758691206543971},
'12h_P2_T4_TimeC3_PIDC2_Survivor': {'DNAJC14': 0.2473118279569893,
'DNAJC30': 0.13333333333333336,
'DNAJC9': 0.12688172043010748},
'24h_P1_T5_TimeC4_PIDC4_Non-Survivor': {'DNAJC14': 0.2416666666666666,
'DNAJC30': 0.13541666666666666,
'DNAJC9': 0.05937499999999994},
'24h_P2_T5_TimeC3_PIDC2_Survivor': {'DNAJC14': 0.23474663908996893,
'DNAJC30': 0.10237849017580147,
'DNAJC9': 0.12616339193381598},
'48h_P1_T6_TimeC3_PIDC1_Non-Survivor': {'DNAJC14': 0.22303664921465965,
'DNAJC30': 0.13821989528795814,
'DNAJC9': 0.13403141361256546},
'48h_P2_T6_TimeC3_PIDC3_Survivor': {'DNAJC14': 0.19415983606557383,
'DNAJC30': 0.11424180327868855,
'DNAJC9': 0.1316598360655738},
'4h_P1_T2_TimeC1_PIDC4_Non-Survivor': {'DNAJC14': 0.2543323139653414,
'DNAJC30': 0.13608562691131498,
'DNAJC9': 0.11162079510703361},
'4h_P2_T2_TimeC2_PIDC1_Survivor': {'DNAJC14': 0.22369765066394287,
'DNAJC30': 0.11542390194075587,
'DNAJC9': 0.09703779366700718},
'8h_P1_T3_TimeC4_PIDC4_Non-Survivor': {'DNAJC14': 0.2451282051282051,
'DNAJC30': 0.11282051282051278,
'DNAJC9': 0.09641025641025636},
'8h_P2_T3_TimeC2_PIDC2_Survivor': {'DNAJC14': 0.22760800842992635,
'DNAJC30': 0.16965226554267654,
'DNAJC9': 0.12750263435194942}}
Expected output (does not have to be exactly like this):
CodePudding user response:
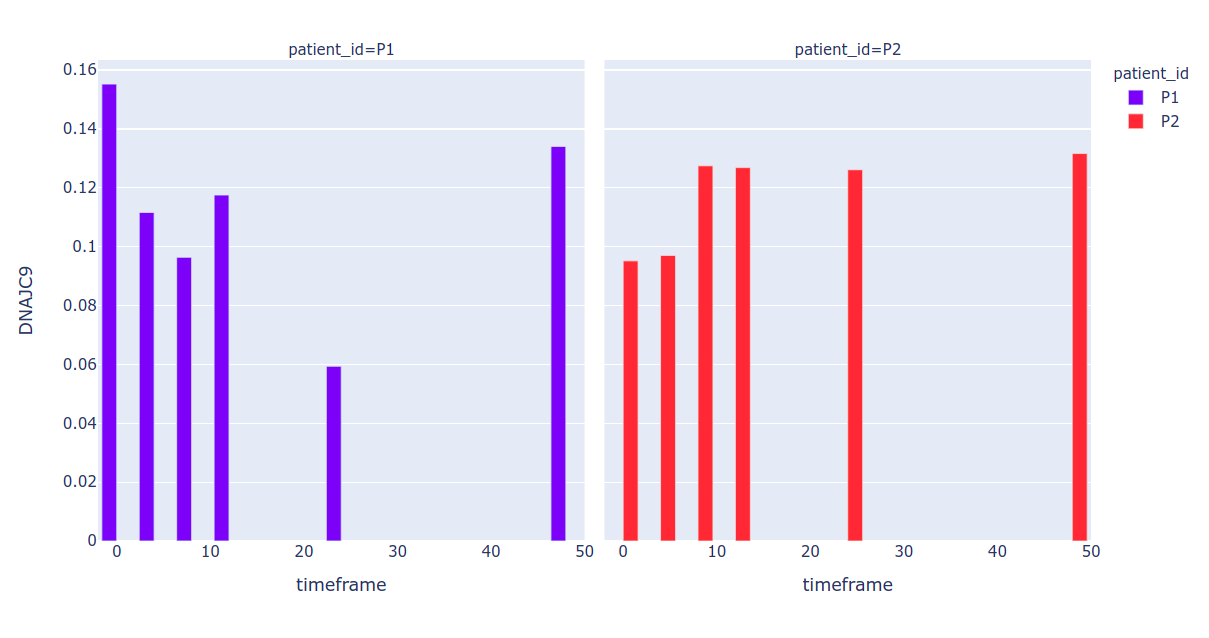
Just for the better view, transpose the dataframe
df = df.T
Derive Patient id and timeframe
df.loc[:, "patient_id"] = df.index.str.split('_').str[1]
Plot the graph
px.bar(df, x="timeframe", y="DNAJC9", color="patient_id", barmode="group",facet_col="patient_id")
Sample output:
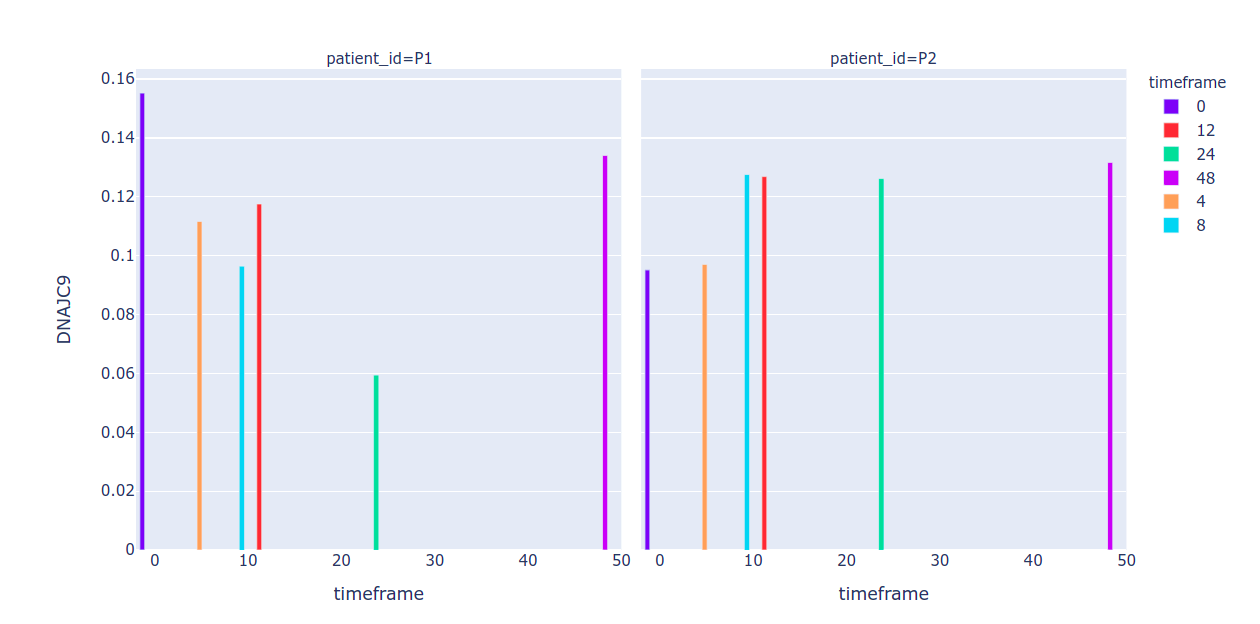
To get separate color bars based on timeframe, keep the timeframe column as object
df.loc[:, "timeframe"] = df.index.str.split('_').str[0].str.replace('h', '')
Sample output:
CodePudding user response:
Add hour and patient:
df = df.T
df['hour'] = df.index.str.split('_').str[0]
df['hour'] = df['hour'].str.replace('h','')
df['patient'] = df.index.str.split('_').str[1]
df = df.astype({'hour':int})
df.sort_values(by=['hour','patient'], inplace=True)
df['hour'] = df['hour'].astype(str) 'h'
df
Plot
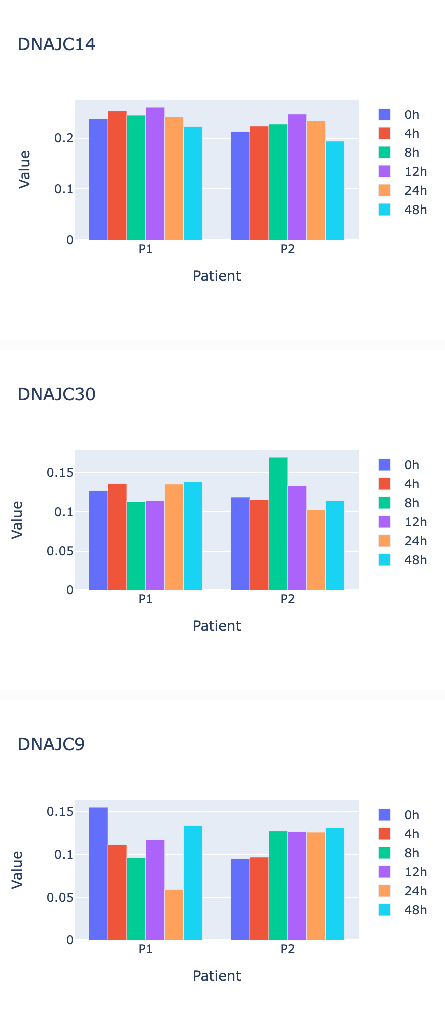
for dnajc in df.columns[:3]:
ob = df[[dnajc, 'hour', 'patient']]
ob.columns = ['value', 'hour', 'patient']
fig = go.Figure(data=[go.Bar(x=ob['patient'], y=ob[ob['hour'] == hr]['value'], name=hr) for hr in ob['hour'].unique()])
fig.update_layout(title=dnajc, xaxis_title='Patient', yaxis_title='Value', width=450, height=320)
fig.show()
Subplot
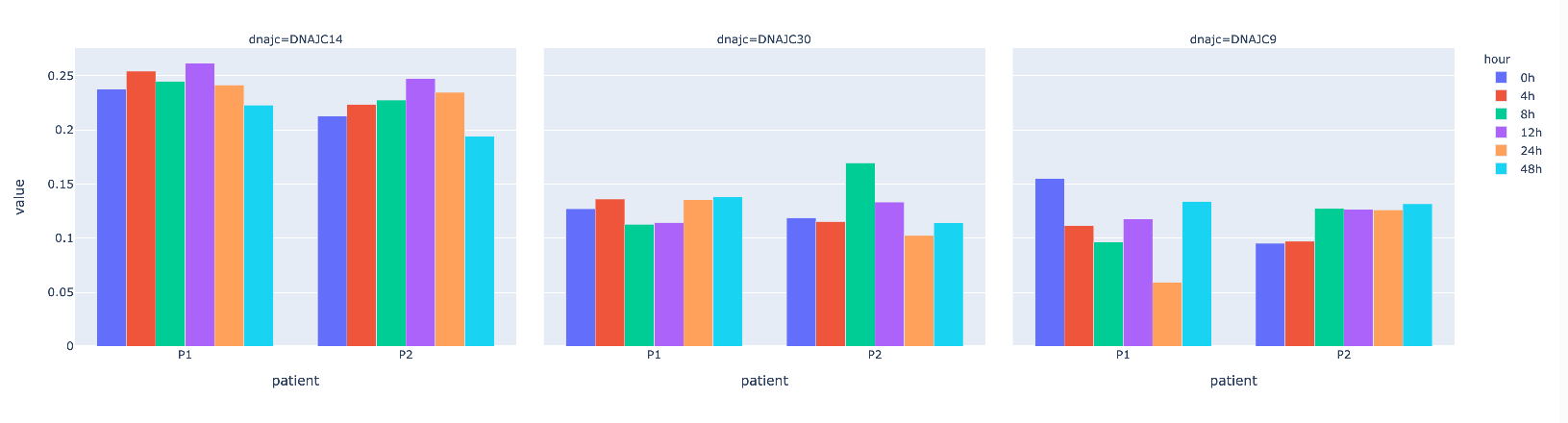
df2 = df.copy()
df2 = df2.melt(id_vars=['hour','patient'], value_vars=df2.columns[:3], var_name='dnajc')
df2
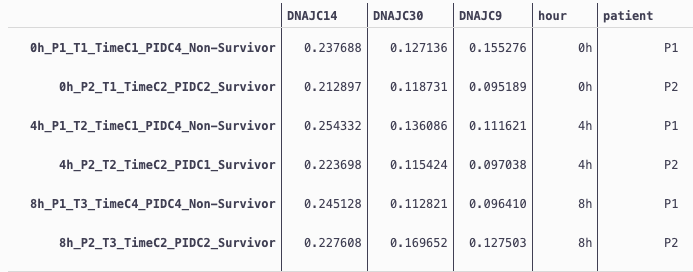
###
hour patient dnajc value
0 0h P1 DNAJC14 0.237688
1 0h P2 DNAJC14 0.212897
2 4h P1 DNAJC14 0.254332
3 4h P2 DNAJC14 0.223698
4 8h P1 DNAJC14 0.245128
5 8h P2 DNAJC14 0.227608
6 12h P1 DNAJC14 0.261759
⋮ ⋮ ⋮ ⋮ ⋮
29 8h P2 DNAJC9 0.127503
30 12h P1 DNAJC9 0.117587
31 12h P2 DNAJC9 0.126882
32 24h P1 DNAJC9 0.059375
33 24h P2 DNAJC9 0.126163
34 48h P1 DNAJC9 0.134031
35 48h P2 DNAJC9 0.131660
Plot
fig_px = px.bar(df2, x='patient', y='value', color='hour', facet_col='dnajc', barmode='group')
fig_px.show()