Problem
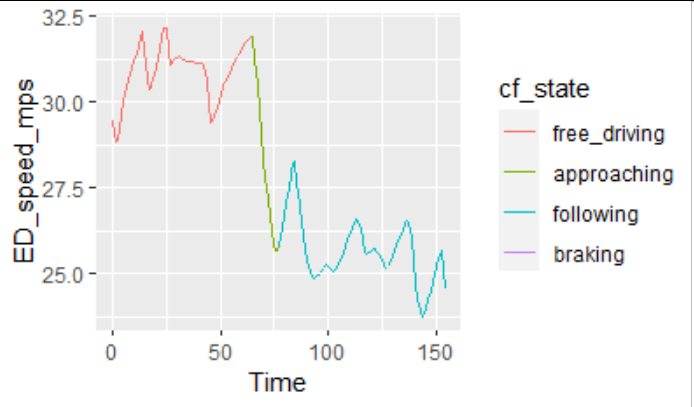
I have observed and simulated speed and states of a car. I have already re-ordered the levels of the states. When I plot the observed speed only, and colour it using the state, I get the following:
ggplot(data = plot_data)
geom_line(aes(x = Time, y = ED_speed_mps, color = cf_state, group = "all"))
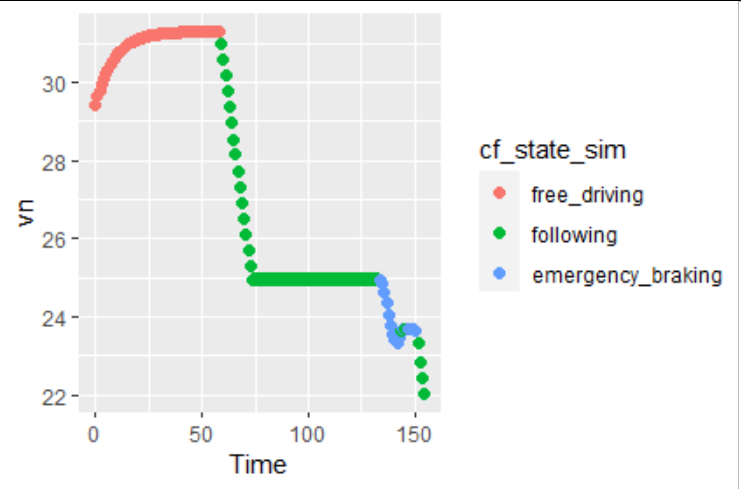
Similarly, simulated speed and states:
ggplot(data = plot_data)
geom_point(aes(x = Time, y = vn, color = cf_state_sim), size = 2)
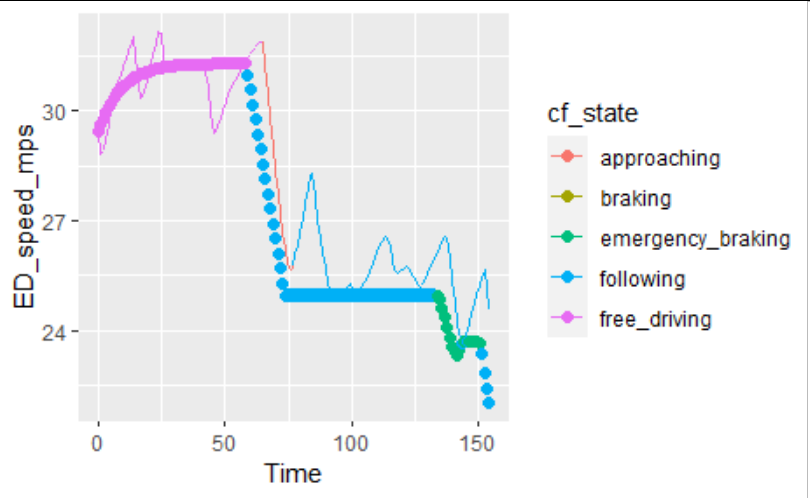
However, when I combine them, the legend order changes and I get a mixed legend:
ggplot(data = plot_data)
geom_line(aes(x = Time, y = ED_speed_mps, color = cf_state, group = "all"))
geom_point(aes(x = Time, y = vn, color = cf_state_sim), size = 2)
How can I separate the legend keys for cf_state (lines) and cf_state_sim (points), while maintaining the order as the original levels of both factor vectors?
Data
Sample data:
plot_data <- structure(list(Time = c(0, 0.98, 1.97, 2.95, 3.93, 4.92, 5.9,
6.88, 7.87, 8.85, 9.83, 10.82, 11.8, 12.78, 13.77, 14.75, 15.73,
16.72, 17.7, 18.68, 19.67, 20.65, 21.63, 22.62, 23.6, 24.58,
25.57, 26.55, 27.53, 28.52, 29.5, 30.48, 31.47, 32.45, 33.43,
34.42, 35.4, 36.38, 37.37, 38.35, 39.33, 40.32, 41.3, 42.28,
43.27, 44.25, 45.23, 46.22, 47.2, 48.18, 49.17, 50.15, 51.13,
52.12, 53.1, 54.08, 55.07, 56.05, 57.03, 58.02, 59, 59.98, 60.97,
61.95, 62.93, 63.92, 64.9, 65.88, 66.87, 67.85, 68.83, 69.82,
70.8, 71.78, 72.77, 73.75, 74.73, 75.72, 76.7, 77.68, 78.67,
79.65, 80.63, 81.62, 82.6, 83.58, 84.57, 85.55, 86.53, 87.52,
88.5, 89.48, 90.47, 91.45, 92.43, 93.42, 94.4, 95.38, 96.37,
97.35, 98.33, 99.32, 100.3, 101.28, 102.27, 103.25, 104.23, 105.22,
106.2, 107.18, 108.17, 109.15, 110.13, 111.12, 112.1, 113.08,
114.07, 115.05, 116.03, 117.02, 118, 118.98, 119.97, 120.95,
121.93, 122.92, 123.9, 124.88, 125.87, 126.85, 127.83, 128.82,
129.8, 130.78, 131.77, 132.75, 133.73, 134.72, 135.7, 136.68,
137.67, 138.65, 139.63, 140.62, 141.6, 142.58, 143.57, 144.55,
145.53, 146.52, 147.5, 148.48, 149.47, 150.45, 151.43, 152.42,
153.4, 154.38), cf_state = structure(c(1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L), .Label = c("free_driving", "approaching",
"following", "braking"), class = "factor"), cf_state_sim = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 3L, 4L, 3L, 4L, 4L, 4L, 4L, 4L, 3L, 3L, 3L, 3L), .Label = c("free_driving",
"approaching", "following", "emergency_braking"), class = "factor"),
ED_speed_mps = c(29.43, 28.81, 28.92, 29.22, 29.62, 30.02,
30.41, 30.68, 30.81, 30.99, 31.2, 31.41, 31.63, 31.84, 32.04,
31.49, 30.72, 30.32, 30.48, 30.7, 30.99, 31.27, 31.54, 31.85,
32.16, 32.15, 31.5, 31.06, 31.16, 31.22, 31.27, 31.33, 31.29,
31.25, 31.23, 31.21, 31.2, 31.19, 31.18, 31.16, 31.15, 31.13,
31.12, 31.1, 30.69, 29.94, 29.41, 29.41, 29.6, 29.8, 30,
30.24, 30.47, 30.6, 30.72, 30.84, 30.97, 31.11, 31.23, 31.36,
31.46, 31.55, 31.64, 31.73, 31.82, 31.9, 31.91, 31.26, 30.52,
29.78, 29.07, 28.36, 27.77, 27.18, 26.67, 26.25, 25.86, 25.6,
25.77, 26.1, 26.34, 26.67, 27.05, 27.48, 27.91, 28.23, 28.3,
27.67, 26.99, 26.51, 26.12, 25.73, 25.35, 25.07, 24.9, 24.85,
24.89, 24.97, 25.03, 25.1, 25.17, 25.27, 25.18, 25.11, 25.06,
25.04, 25.21, 25.34, 25.47, 25.61, 25.79, 25.98, 26.16, 26.32,
26.45, 26.58, 26.53, 26.32, 25.97, 25.65, 25.58, 25.61, 25.66,
25.72, 25.74, 25.64, 25.52, 25.4, 25.28, 25.17, 25.23, 25.38,
25.54, 25.69, 25.84, 25.98, 26.13, 26.27, 26.42, 26.55, 26.51,
26.01, 25.37, 24.74, 24.23, 23.91, 23.75, 23.79, 24, 24.24,
24.48, 24.73, 24.97, 25.21, 25.44, 25.65, 25.16, 24.54),
vn = c(29.43, 29.63, 29.8, 29.96, 30.1, 30.22, 30.34, 30.44,
30.53, 30.61, 30.68, 30.75, 30.81, 30.86, 30.91, 30.95, 30.98,
31.02, 31.05, 31.08, 31.1, 31.12, 31.14, 31.16, 31.17, 31.19,
31.2, 31.21, 31.22, 31.23, 31.24, 31.25, 31.25, 31.26, 31.26,
31.27, 31.27, 31.28, 31.28, 31.28, 31.28, 31.29, 31.29, 31.29,
31.29, 31.29, 31.3, 31.3, 31.3, 31.3, 31.3, 31.3, 31.3, 31.3,
31.3, 31.3, 31.3, 31.3, 31.3, 31.3, 30.99, 30.58, 30.18,
29.77, 29.36, 28.96, 28.55, 28.15, 27.74, 27.33, 26.93, 26.52,
26.11, 25.71, 25.3, 24.96, 24.97, 24.96, 24.97, 24.96, 24.97,
24.96, 24.97, 24.96, 24.97, 24.96, 24.97, 24.96, 24.97, 24.96,
24.97, 24.96, 24.97, 24.96, 24.97, 24.96, 24.97, 24.96, 24.97,
24.96, 24.97, 24.96, 24.97, 24.96, 24.97, 24.96, 24.97, 24.96,
24.97, 24.96, 24.97, 24.96, 24.97, 24.96, 24.97, 24.96, 24.97,
24.96, 24.97, 24.96, 24.97, 24.96, 24.97, 24.96, 24.97, 24.96,
24.97, 24.96, 24.97, 24.96, 24.97, 24.96, 24.97, 24.96, 24.97,
24.96, 24.96, 24.84, 24.63, 24.36, 24.06, 23.79, 23.57, 23.41,
23.32, 23.48, 23.65, 23.68, 23.7, 23.7, 23.7, 23.71, 23.71,
23.64, 23.35, 22.82, 22.42, 22.01)), row.names = c(NA, -158L
), class = c("tbl_df", "tbl", "data.frame"))
CodePudding user response:
I don't think {ggplot2} will let you split legends if it thinks they should be merged. However the {ggnewscale} package can add this functionality. You can provide named vector of colors to ggplot2::scale_color_manual(values = ...) to separately control the color mapping for each scale. And with new_scale_color() you'll get to use scale_color_manual() twice and provide the color scale you like to each separately.
The {relayer} package should have similar functionality to allow this but I wasn't able to get it working on your example for some reason.
## split legend with {ggnewscale}
library(ggplot2)
library(ggnewscale)
ggplot(data = plot_data, aes(x = Time))
geom_line(aes(y = ED_speed_mps, color = cf_state, group = "all"))
ggnewscale::new_scale_color()
geom_point(aes(y = vn, color = cf_state_sim), size = 2)
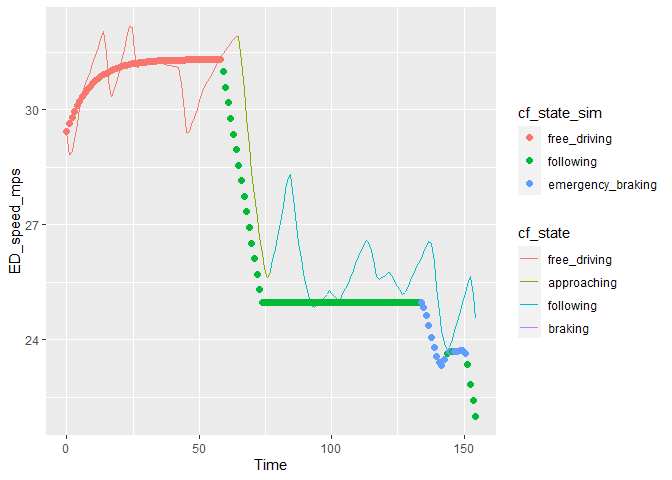
Created on 2022-07-27 by the reprex package (v2.0.1)