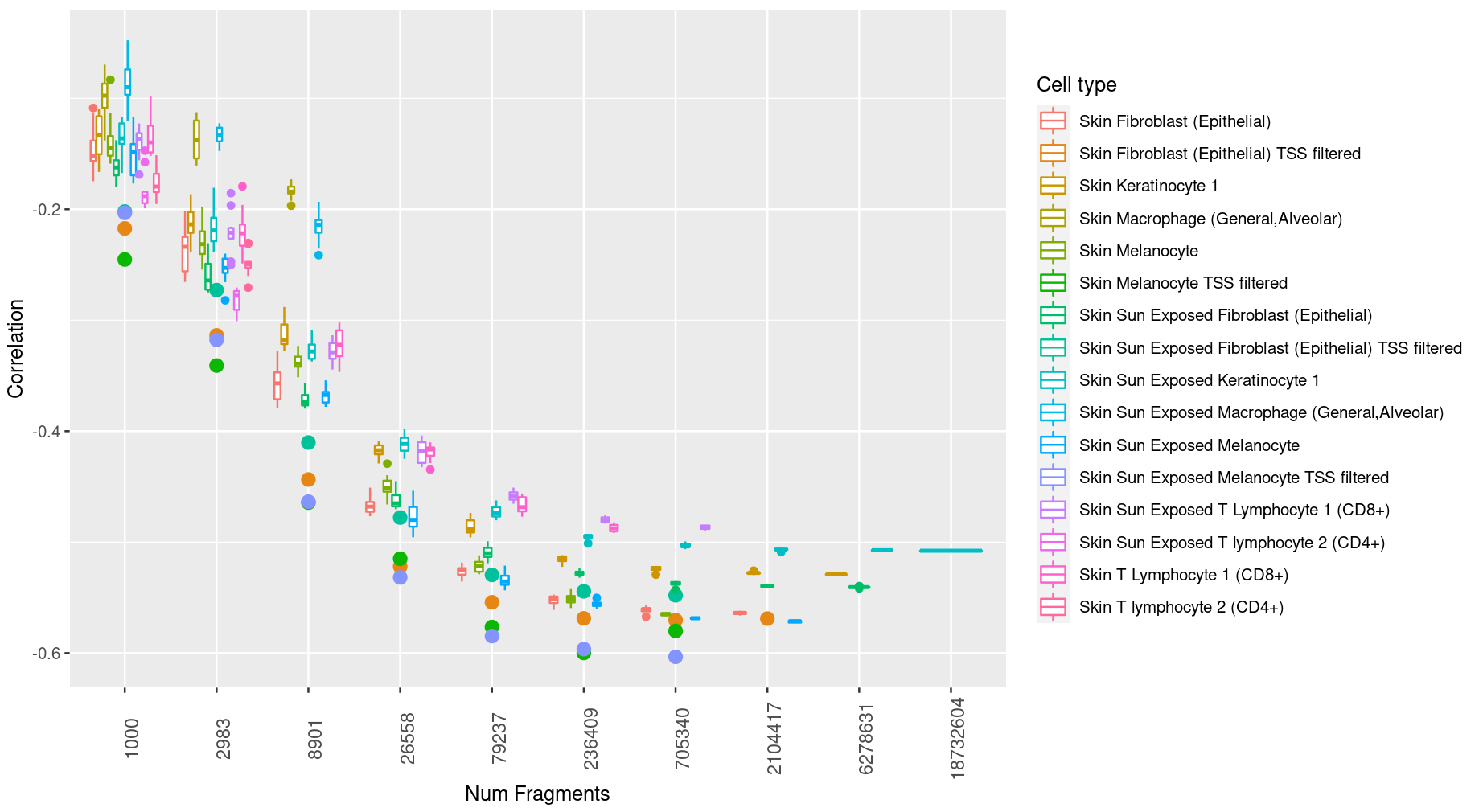
I'm using two geoms from ggplot, geom_point and geom_boxplot, to create the plot shown below. Note that the legend for the points from geom_point is incorrect, since the points are being treated as boxplots (in the legend, anything that has the string "TSS filtered" appended at the end is supposed to be a point instead of a boxplot). I'm not exactly sure how to fix this.
I've included the output from dput for df (which has the data for the points) and and an abbreviated output from dput for correlations_long with only Skin Sun Exposed Melanocyte at the end of this post (correlations_long has the data for the boxplots).
ggplot()
geom_point(data=df, aes(x=factor(name, levels=unique(name)),
y=value, color=cell_type), size=3)
geom_boxplot(data=correlations_long,
aes(x=factor(num_fragments, levels=unique(num_fragments)),
y=correlation, color=cell_type))
theme(axis.text.x=element_text(size=10, angle=90))
xlab("Num Fragments")
ylab("Correlation")
labs(color="Cell type")
scale_x_discrete(limits = as.factor(sort(as.integer(
unique(correlations_long["num_fragments"]) %>% pull))))
dput output for df:
structure(list(name = c("1000", "2983", "8901", "26558", "79237",
"236409", "705340", "2104417", "1000", "2983", "8901", "26558",
"79237", "236409", "705340", "2104417", "1000", "2983", "8901",
"26558", "79237", "236409", "705340", "1000", "2983", "8901",
"26558", "79237", "236409", "705340"), value = c(-0.217233727868593,
-0.313682277356269, -0.443536645598551, -0.521959275706954, -0.554173539213182,
-0.568676467365853, -0.570234215267721, -0.568882712628573, -0.245220070304608,
-0.340830882171965, -0.464351593030875, -0.514900990218652, -0.576472067464575,
-0.59987452898425, -0.58002528101756, NA, -0.202054800162606,
-0.272836084111954, -0.410191142797998, -0.477791235306681, -0.529504522998714,
-0.544302595354142, -0.547864149527051, -0.203245452534301, -0.317663927450409,
-0.463555979106284, -0.531779779812837, -0.584611371556314, -0.596363138503893,
-0.603323902370038), cell_type = c("Skin Fibroblast (Epithelial) TSS filtered",
"Skin Fibroblast (Epithelial) TSS filtered", "Skin Fibroblast (Epithelial) TSS filtered",
"Skin Fibroblast (Epithelial) TSS filtered", "Skin Fibroblast (Epithelial) TSS filtered",
"Skin Fibroblast (Epithelial) TSS filtered", "Skin Fibroblast (Epithelial) TSS filtered",
"Skin Fibroblast (Epithelial) TSS filtered", "Skin Melanocyte TSS filtered",
"Skin Melanocyte TSS filtered", "Skin Melanocyte TSS filtered",
"Skin Melanocyte TSS filtered", "Skin Melanocyte TSS filtered",
"Skin Melanocyte TSS filtered", "Skin Melanocyte TSS filtered",
"Skin Melanocyte TSS filtered", "Skin Sun Exposed Fibroblast (Epithelial) TSS filtered",
"Skin Sun Exposed Fibroblast (Epithelial) TSS filtered", "Skin Sun Exposed Fibroblast (Epithelial) TSS filtered",
"Skin Sun Exposed Fibroblast (Epithelial) TSS filtered", "Skin Sun Exposed Fibroblast (Epithelial) TSS filtered",
"Skin Sun Exposed Fibroblast (Epithelial) TSS filtered", "Skin Sun Exposed Fibroblast (Epithelial) TSS filtered",
"Skin Sun Exposed Melanocyte TSS filtered", "Skin Sun Exposed Melanocyte TSS filtered",
"Skin Sun Exposed Melanocyte TSS filtered", "Skin Sun Exposed Melanocyte TSS filtered",
"Skin Sun Exposed Melanocyte TSS filtered", "Skin Sun Exposed Melanocyte TSS filtered",
"Skin Sun Exposed Melanocyte TSS filtered")), row.names = c(NA,
-30L), class = c("data.table", "data.frame"))
dput output for correlations_long:
structure(list(num_fragments = c("1000", "2983", "8901", "26558",
"79237", "236409", "705340", "2104417", "1000", "2983", "8901",
"26558", "79237", "236409", "705340", "2104417", "1000", "2983",
"8901", "26558", "79237", "236409", "705340", "2104417", "1000",
"2983", "8901", "26558", "79237", "236409", "705340", "2104417",
"1000", "2983", "8901", "26558", "79237", "236409", "705340",
"2104417", "1000", "2983", "8901", "26558", "79237", "236409",
"705340", "2104417", "1000", "2983", "8901", "26558", "79237",
"236409", "705340", "2104417", "1000", "2983", "8901", "26558",
"79237", "236409", "705340", "2104417", "1000", "2983", "8901",
"26558", "79237", "236409", "705340", "2104417", "1000", "2983",
"8901", "26558", "79237", "236409", "705340", "2104417"), correlation = c(-0.142552912633417,
-0.282063642684633, -0.358365031342374, -0.476429739615196, -0.537715257634154,
-0.556507228835608, -0.569290141585588, -0.572632853618199, -0.176729246694482,
-0.24002202483771, -0.364372461559598, -0.472537637457574, -0.521181023255881,
-0.55984376716404, -0.568700874753408, -0.571018314483162, -0.140246012715342,
-0.265674247705073, -0.375020572402195, -0.466325284307382, -0.543448139139435,
-0.554991371026006, -0.567841698567904, -0.572626483191379, -0.1626413145619,
-0.243304716053789, -0.371378032491436, -0.483941606311232, -0.530845237866024,
-0.554468643820047, -0.568143694079138, -0.570862620697964, -0.116564401636032,
-0.255281310962979, -0.377464344414864, -0.48674764840038, -0.534623306115981,
-0.559268330681496, -0.568358687051924, -0.572804016053457, -0.150598835600339,
-0.250340179755359, -0.378018309624652, -0.483361453380124, -0.539940888020042,
-0.550240697380326, -0.568095658044177, -0.571273467002167, -0.140801591191654,
-0.243907992257946, -0.365389408139248, -0.466829280870502, -0.53860508515168,
-0.554893906404059, -0.567088355995875, -0.571123984164459, -0.175643182028142,
-0.257385314673002, -0.354177687837023, -0.494490415619058, -0.528805416429225,
-0.556373311038017, -0.569306986486968, -0.572012337412355, -0.171542682526591,
-0.246289070052161, -0.366099952822677, -0.453690709343421, -0.530025481091536,
-0.551777830306486, -0.568937084190358, -0.570650095061955, -0.146629007977338,
-0.257264392122815, -0.36851473188057, -0.495616527451394, -0.536316439462659,
-0.557762199945624, -0.569946797833086, -0.57003184524515), cell_type = c("Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte", "Skin Sun Exposed Melanocyte",
"Skin Sun Exposed Melanocyte")), row.names = c(NA, -80L), class = c("tbl_df",
"tbl", "data.frame"))
CodePudding user response:
You've got a few options. One way is to specify which key glyph to use depending on the color (i.e. if it's red, yellow, green or purple use a dot symbol, and if it's blue use a boxplot symbol; https://ggplot2.tidyverse.org/reference/draw_key.html), e.g.
library(tidyverse)
# To get the hex codes for the default ggplot2 colorscheme:
# scales::show_col(scales::hue_pal()(5))
# [1] "#F8766D" "#A3A500" "#00BF7D" "#00B0F6" "#E76BF3"
# Create a custom key_glyph function:
draw_key_cust <- function(data, params, size) {
if (data$colour %in% c("#F8766D", "#A3A500", "#00BF7D", "#E76BF3")) {
draw_key_point(data, params, size)
} else if (data$colour == "#00B0F6") {
draw_key_boxplot(data, params, size)
} else {
draw_key_point(data, params, size)
}
}
ggplot()
geom_point(data=df, aes(x=factor(name, levels=unique(name)),
y=value, color=cell_type), size=3)
geom_boxplot(data=correlations_long,
aes(x=factor(num_fragments, levels=unique(num_fragments)),
y=correlation, color=cell_type),
key_glyph = "cust")
theme(axis.text.x=element_text(size=10, angle=90))
xlab("Num Fragments")
ylab("Correlation")
labs(color="Cell type")
scale_x_discrete(limits = as.factor(sort(as.integer(
unique(correlations_long["num_fragments"]) %>% pull))))
#> Warning: Removed 1 rows containing missing values (geom_point).
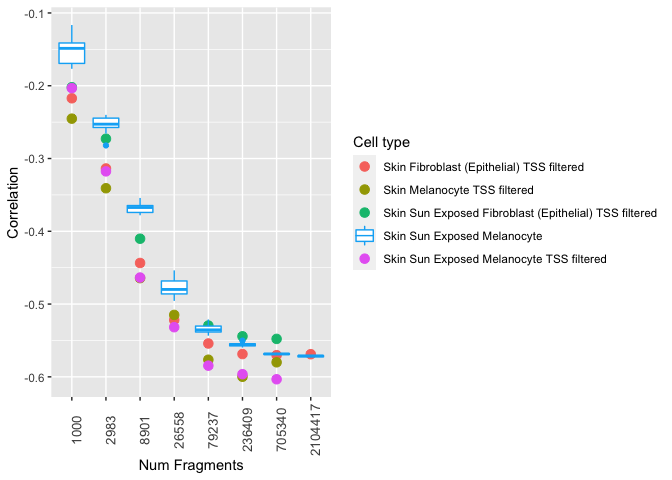
Another way is to split up the legend into two (i.e. one for color, one for fill) and this specifies the symbols accordingly:
library(tidyverse)
ggplot()
geom_point(data=df, aes(x=factor(name, levels=unique(name)),
y=value, fill=cell_type), shape = 21, size=3)
geom_boxplot(data=correlations_long,
aes(x=factor(num_fragments, levels=unique(num_fragments)),
y=correlation, color=cell_type))
theme(axis.text.x=element_text(size=10, angle=90))
xlab("Num Fragments")
ylab("Correlation")
labs(color="Cell type", fill = "Cell type")
scale_x_discrete(limits = as.factor(sort(as.integer(
unique(correlations_long["num_fragments"]) %>% pull))))
#> Warning: Removed 1 rows containing missing values (geom_point).
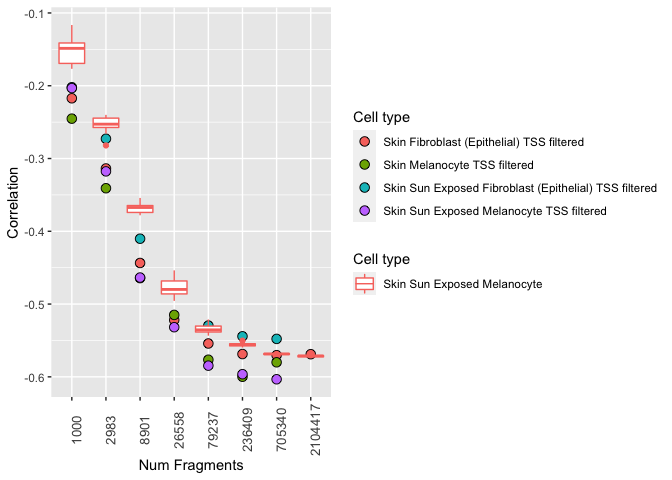
Or, you can stick with the combined legend and combine the key glyphs (i.e. a boxplot and a dot for each):
ggplot()
geom_boxplot(data=correlations_long,
aes(x=factor(num_fragments, levels=unique(num_fragments)),
y=correlation, color=cell_type),
key_glyph = "boxplot")
geom_point(data=df, aes(x=factor(name, levels=unique(name)),
y=value, color=cell_type), size=3,
key_glyph = "point")
theme(axis.text.x=element_text(size=10, angle=90))
xlab("Num Fragments")
ylab("Correlation")
labs(color="Cell type", fill = "Cell type")
scale_x_discrete(limits = as.factor(sort(as.integer(
unique(correlations_long["num_fragments"]) %>% pull))))
#> Warning: Removed 1 rows containing missing values.
Created on 2022-08-23 by the reprex package (v2.0.1)
In my opinion the first option is the best, but it's also the most complicated; if you need help implementing it, edit your question then tag me in a comment and I'll help you troubleshoot.